Genome-wide Analysis and Identification of Phosphoinositide-specific Phospholipase C Gene Family in Poplar (Populus trichocarpa)
-
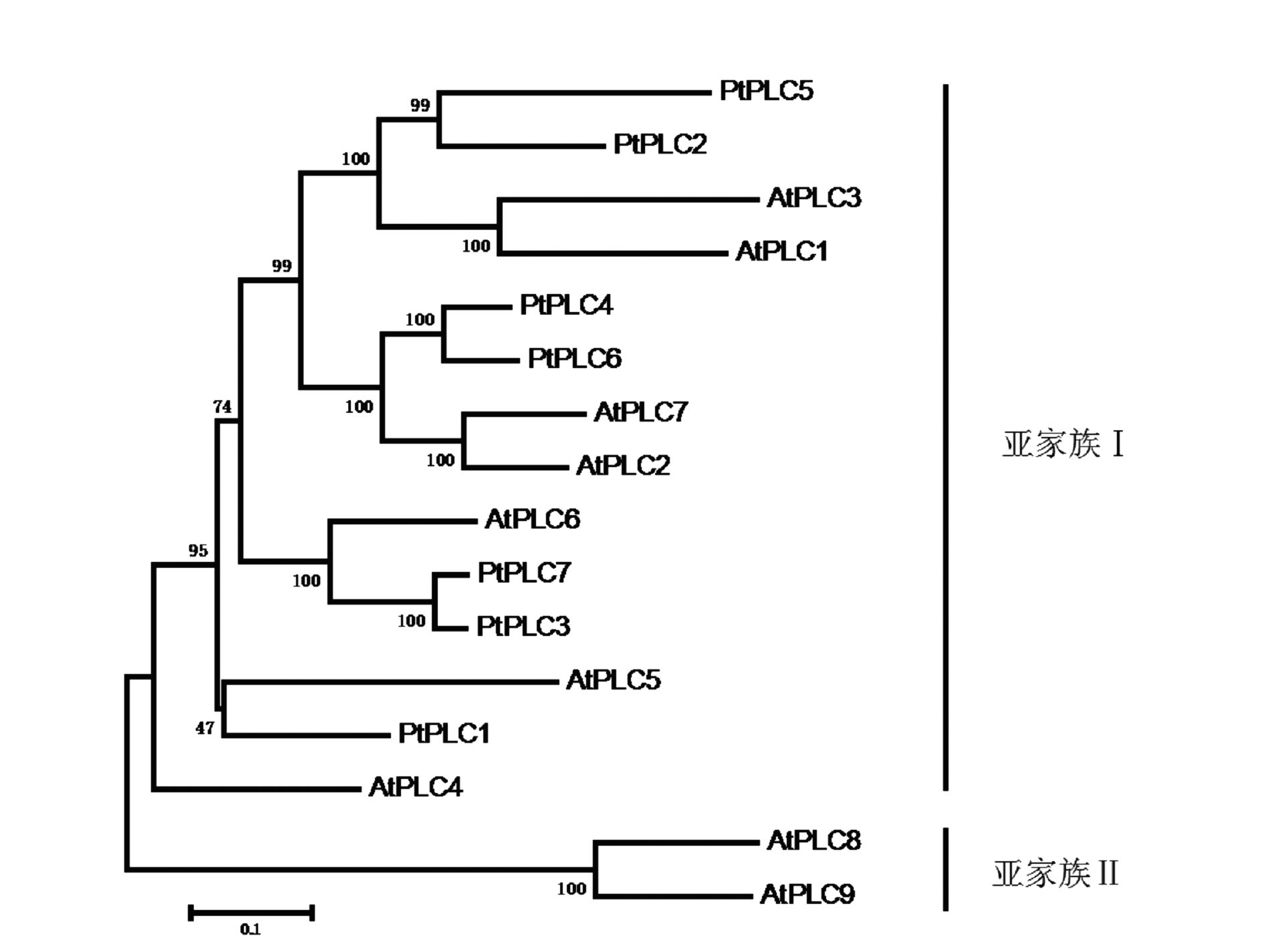
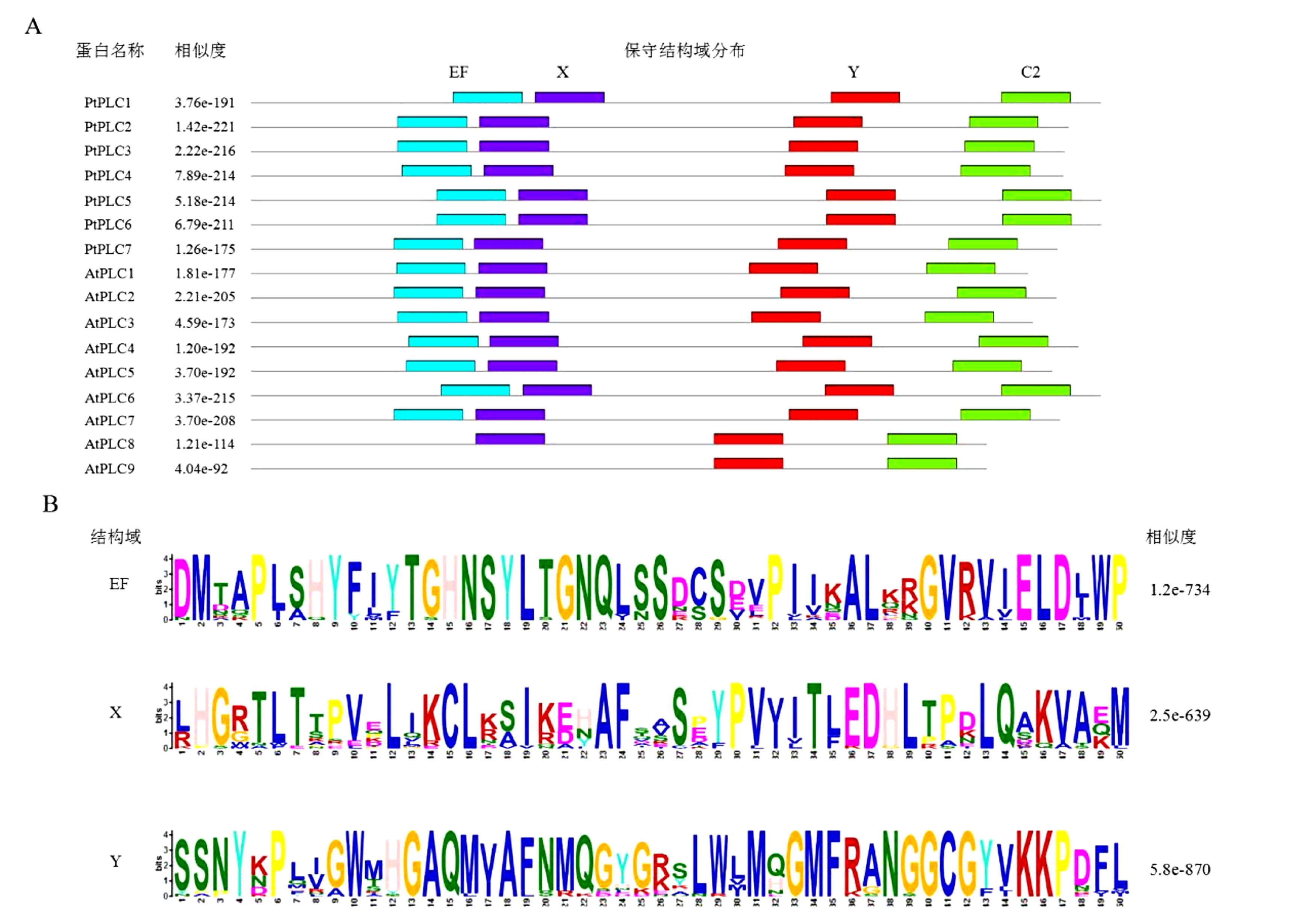
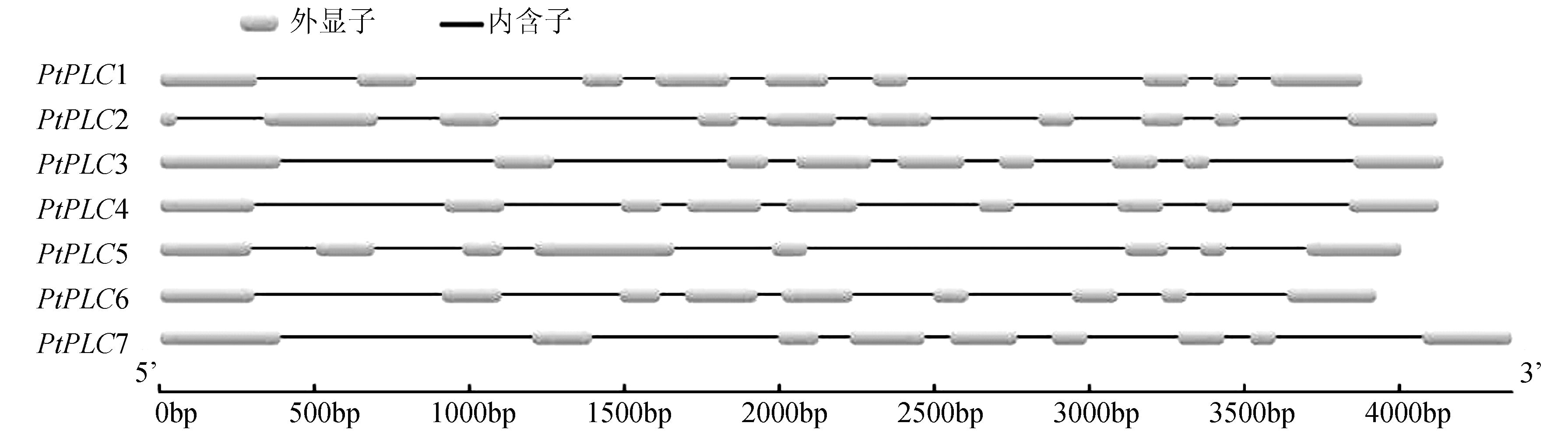
摘要: 为了对已测序杨树Populus trichocarpa Torr. & Gray中磷酸肌醇特异性磷脂酶C(phosphoinositide-specific phospholipase C,PI-PLC)家族基因进行系统分析,利用杨树基因组数据库,通过生物信息学手段,鉴定杨树PI-PLC 家族基因的基因结构、染色体定位和编码蛋白,通过蛋白序列比对进行进化和分类分析。结果表明,杨树中含有7个PI-PLC家族基因,含有8~10个外显子,分布于杨树的4条染色体上。MEME和Pfam保守结构域分析显示,杨树PI-PLC蛋白均含有4个保守的EF、X、Y和C2结构域。蛋白质进化树分析结果表明PI-PLC可分为2个亚家族。
-
关键词:
- 磷酸肌醇特异性磷脂酶C /
- 杨树 /
- 进化分析 /
- 基因家族
Abstract: Phosphoinositide-specific phospholipase C (PI-PLC) is a conserved key enzyme in phosphoinositide signaling for animals and plants. It plays an important role in growth and development, as well as various abiotic and biotic stresses in the organisms. Based upon the existing database and bioinformatics methodologies, PI-PLC family gene structures and their positions on chromosome and duplication for poplar (Populus trichocarpa Torr. & Gray) (Pt) were obtained. PI-PLC proteins were classified according to their phylogenetic relationship. The results showed that 7 PtPLC genes existed in the poplar genome on 4 chromosomes. Their multiple alignments and motif displays indicated that all of the PtPLC proteins contained 4 conserved domains (i.e., EF, X, Y, and C2). Phylogenetic analysis on the PtPLC family classified them into two groups. The information would be the basis for a study on the functions of PtPI-PLC gene family.-
Key words:
- PI-PLC /
- Populus trichocarpa /
- phylogeny analysis /
- gene family
-
表 1 杨树PI-PLC家族基因的基本特征
Table 1. Characteristics of PI-PLC genes of poplar
基因名称 定位名称 转录名称 染色体定位
染色体开放阅读
框位置蛋白
/bp外显子个数
/aaPtPLC1 Potri.001G252100 1G252100.1 Chr01:26180768..26185026 forward 1761 586 9 PtPLC2 Potri.001G252300 1G252300.1 Chr01:26191421..26195808 forward 1842 613 10 PtPLC3 Potri.008G068300 8G068300.1 Chr08:4159193..4163941 forward 1845 614 9 PtPLC4 Potri.008G068400 8G068400.1 Chr08:4165486..4170103 forward 1773 590 9 PtPLC5 Potri.009G046600 9G046600.1 Chr09:5204380..5208632 forward 1749 582 8 PtPLC6 Potri.010G188800 10G188800.1 Chr10:18467139..18471555 reverse 1764 587 9 PtPLC7 Potri.010G188900 10G188900.1 Chr10:18476727..18481689 reverse 1845 614 9 表 2 杨树基因组中PI-PLC 蛋白氨基酸相似性分析
Table 2. Amino acid seque nce similarity analysis of the OtPLC proteins
PI-PLC 家族 PtPLC2 PtPLC3 PtPLC4 PtPLC5 PtPLC6 PtPLC7 PtPLC1 57.37 65.75 64.64 52.12 65.68 65.92 PtPLC2 57.50 62.17 67.55 62.11 57.17 PtPLC3 63.75 52.20 62.52 92.67 PtPLC4 54.97 88.93 63.92 PtPLC5 54.91 51.67 PtPLC6 63.39 表 3 杨树PtPLC1、4、6和7不同转录本的基本特征
Table 3. Characteristics of PtPLC1,4,6,and 7 genes
基因名称 定位名称 转录名称 别名 染色体定位
染色体开放阅读
框位置蛋白
/bp外显子
个数/aaPtPLC1 Potri.001G252100 Potri.001G252100.1 POPTR_0001s25910/.v2.2/.1.v2.2 Chr01:26180768..26185026 + 1761 586 9 PtPLC1 Potri.001G252100 Potri.001G252100.2 POPTR_0001s25910/.v2.2 Chr01:26180768..26185025 + 1107 368 5 PtPLC1 Potri.001G252100 Potri.001G252100.3 POPTR_0001s25910/.v2.2 Chr01:26180768..26185025 + 1467 488 7 PtPLC1 Potri.001G252100 Potri.001G252100.4 POPTR_0001s25910/.v2.2 Chr01:26180768..26183407 + 1227 408 6 PtPLC4 Potri.008G068400 Potri.008G068400.1 POPTR_0008s06840/.v2.2/.1.v2.2 Chr08:4165486..4170103 + 1773 590 9 PtPLC4 Potri.008G068400 Potri.008G068400.2 POPTR_0008s06840/.v2.2 Chr08:4165517..4168012 + 1167 388 5 PtPLC6 Potri.010G188800 Potri.010G188800.1 POPTR_0010s19610/.v2.2/.1.v2.2 Chr10:18467139..18471555- 1764 587 9 PtPLC6 Potri.010G188800 Potri.010G188800.2 POPTR_0010s19610/.v2.2 Chr10:18467139..18471555- 1557 518 9 PtPLC6 Potri.010G188800 Potri.010G188800.3 POPTR_0010s19610/.v2.2 Chr10:18467139..18471555- 1518 505 8 PtPLC6 Potri.010G188800 Potri.010G188800.4 POPTR_0010s19610/.v2.2 Chr10:18467139..18471555- 1407 468 8 PtPLC6 Potri.010G188800 Potri.010G188800.5 POPTR_0010s19610/.v2.2 Chr10:18467139..18471555- 1122 373 6 PtPLC6 Potri.010G188800 Potri.010G188800.6 POPTR_0010s19610/.v2.2 Chr10:18467139..18470622- 1443 480 8 PtPLC7 Potri.010G188900 Potri.010G188900.1 POPTR_0010s19620/.v2.2 Chr10:18476727..18481689- 1845 614 9 PtPLC7 Potri.010G188900 Potri.010G188900.2 POPTR_0010s19620/.v2.2/.1.v2.2 Chr10:18476727..18481689- 1788 595 9 PtPLC7 Potri.010G188900 Potri.010G188900.3 POPTR_0010s19620/.v2.2 Chr10:18479368..18481446- 339 112 2 -
[1] RUPWATE S D,RAJASEKHARAN R.Plant phosphoinositide-specific phospholipase C:an insight[J]. Plant Signa Behav, 2012, 7(10):1281-1283. doi: 10.4161/psb.21436 [2] MUNNIK T, TESTERINK C. Plant phospholipid signaling:"in a nutshell"[J]. Lipid Res,2009, 50(S1):S260-S265. http://cn.bing.com/academic/profile?id=ba3acc5cf49fee0c1efadf719bca6adc&encoded=0&v=paper_preview&mkt=zh-cn [3] WANG X, DEVAIAH S P, ZHANG W, et al. Signaling functions of phosphatidic acid[J]. Prog Lipid Res, 2006, 45(3):250-278. doi: 10.1016/j.plipres.2006.01.005 [4] TAKAHASHI S, KATAGIRI T, HIRAYAMA T, et al. Hyperosmotic stress induces a rapid and transient increase in inositol 1, 4, 5-trisphosphate independent of abscisic acid in Arabidopsis cell culture[J]. Plant Cell Physiol, 2001,42(2):214-222. doi: 10.1093/pcp/pce028 [5] KANEHARA K, YU C Y, CHO Y, et al. Arabidopsis AtPLC2 is a primary phosphoinositide-specific phospholipase C in phosphoinositide metabolism and the endoplasmic reticulum stress response[J]. PLoS Genet, 2015,11(9):e1005511. doi: 10.1371/journal.pgen.1005511 [6] LI L, HE Y, WANG Y, et al. Arabidopsis PLC2 is involved in auxin-modulated reproductive development[J]. Plant J, 2015, 84(3):504-515. doi: 10.1111/tpj.2015.84.issue-3 [7] ZHENG S Z, LIU Y L, LI B, et al. Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance in Arabidopsis[J]. Plant J, 2012, 69(4):689-700. doi: 10.1111/tpj.2012.69.issue-4 [8] GAO K, LIU Y L, LI B, et al. Arabidopsis thaliana phosphoinositide-specific phospholipase C isoform 3(AtPLC3) and AtPLC9 have an additive effect on thermotolerance[J]. Plant Cell Physiol, 2014, 55(11):1873-1883. doi: 10.1093/pcp/pcu116 [9] ZHANG J, XIA K, YANG Y, et al. Overexpression of Arabidopsis phosphoinositide-specific phospholipase C5 induces leaf senescence[J]. Plant Cell Tiss Organ Cult, 2015, 120(2):585-595. doi: 10.1007/s11240-014-0625-y [10] TUSKAN G A, DIFAZIO S, JANSSON S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray)[J]. Science, 2006,313(5793):1596-1604. doi: 10.1126/science.1128691 [11] LARKIN M A, BLACKSHIELDS G, BROWN N P, et al. Clustal W and clustal X version 2.0[J]. Bioinformatics, 2007, 23(21):2947-2948. doi: 10.1093/bioinformatics/btm404 [12] TAMURA K, STECHER G, PETERSON D, et al. MEGA6:molecular evolutionary genetics analysis version 6.0[J]. Mol Biol Evol, 2013, 30(12):2725-2729. doi: 10.1093/molbev/mst197 [13] BAILEY T L, BODEN M, BUSKE F A, et al. MEME SUITE:Tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue):W202-208. http://cn.bing.com/academic/profile?id=930f9869f15cf1dfd5b422c191ce03ed&encoded=0&v=paper_preview&mkt=zh-cn [14] FINN R D, COGGILL P, EBERHARDT R Y, et al. The Pfam protein families database:towards a more sustainable future[J]. Nucleic Acids Res, 2016, 44(Database issue):D279-D285. http://cn.bing.com/academic/profile?id=e0bb014b507fe8f798a3b45993811773&encoded=0&v=paper_preview&mkt=zh-cn [15] TRIPATHY M K, TYAGI W, GOSWAMI M, et al. Characterization and functional validation of tobacco PLCδ for abiotic stress tolerance[J]. Plant Mol Biol Rep, 2012,30(2):488-497. doi: 10.1007/s11105-011-0360-z [16] WANG C R, YANG A F, YUE G D, et al. Enhanced expression of phospholipase C 1(ZmPLC1) improves drought tolerance in transgenic maize[J]. Planta, 2008, 227(5):1127-1140. doi: 10.1007/s00425-007-0686-9 [17] PAOLO G D, CAMILLI P D. Phosphoinositides in cell regulation and membrane dynamics[J]. Nature, 2006, 443(7112):651-657. doi: 10.1038/nature05185 [18] COURSOL S, GIGLIOLI-GUIVARC'H N, VIDAL J, et al. An increase in phosphoinositide-specific phospholipase C activity precedes induction of C4 phosphoenolpyruvate carboxylase phosphorylation in illuminated and NH4Cl-treated protoplasts from Digitaria sanguinalis[J]. Plant J, 2000, 23(4):497-506. doi: 10.1046/j.1365-313x.2000.00819.x [19] SINGH A, KANWAR P, PANDEY A,et al. Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice[J]. PLoS ONE, 2013, 8(4):e62494. doi: 10.1371/journal.pone.0062494 [20] HIRAYAMA T, OHTO C, MIZOGUCHI T, et al. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 1995, 92(1):3903-3907. http://cn.bing.com/academic/profile?id=e7b50273004707669b83a8988d14b2fb&encoded=0&v=paper_preview&mkt=zh-cn [21] SANCHEZ J P, CHUA N H. Arabidopsis PLC1 is required for secondary responses to abscisic acid signals[J]. Plant Cell, 2001,13(5):1143-1154. doi: 10.1105/tpc.13.5.1143 [22] XU X, CAO Z, LIU G, et al. Cloning and expression of AtPLC6, a gene encoding a phosphatidylinositol-specific Arabidopsis thaliana[J]. Chinese Sci Bull, 2004, 49(6):567-573. doi: 10.1360/03wc0514 [23] ZHANG J, ZHANG Z, ZHU D, et al. Expression and initial characterization of a Phosphoinositide-specific phospholipase C from Populus tomentosa[J]. J Plant Biochem Biotech, 2015, 24(3):338-346. doi: 10.1007/s13562-014-0279-1 -







 下载:
下载: