The Dynamic Change of Gene Expression from Pinus massoniana in Response to Bursaphelenchus xylophilus Infestation
-
摘要: 松材线虫病是一种严重危害马尾松生长的流行性病害,可导致马尾松枯萎死亡。为揭示该过程中基因的表达变化行为,本研究利用荧光定量PCR技术检测了松材线虫侵染不同天数下的马尾松较其对照样本中病原识别、抗逆调节、次生代谢、解毒作用及生长素响应等相关基因的表达变化。结果显示,除了与病原识别相关的CC-NBS-LRR抗性蛋白基因的表达随侵染天数的增加而增强之外,其他基因则在侵染2 d的马尾松样本中的表达水平最高,且明显高于未受侵染的对照马尾松样本,随后在侵染3天的马尾松中的表达又低于对照样本。此外,黄酮-3-羟化酶和细胞色素P450基因的表达随着侵染虫量的增加呈先上调后下调的变化方式。通过本研究初步揭示了马尾松响应松材线虫侵染的基因表达变化模式。Abstract: Pine wilt disease (PWD) is a devastating disease, which affects on the growth of masson pine (Pinus massoniana), often leading to withering and death. To reveal the change of gene expression pattern on this process, the gene dynamic expression from the P. massoniana infestation with nematode (Bursaphelenchus xylophilus) for 1, 2, and 3 days was studied. The genes, which were relevant to pathogen recognize, stress regulation, secondary metabolism, detoxification, and auxin-response were concerned. The results showed that most of the tested genes were presented highest expression level on the P. massoniana with infestation for two days, it was then down-regulated on these groups with 3-day infestation. Besides, the gene expression of CC-NBS-LRR resistance protein was continually increased in the treated P. massoniana compared with the corresponding control group. Additionally, genes expression of flavonoid 3-hydroxylase and cytochrome P450 were firstly up-regulated and then down-regulated on the P. massoniana respectively infected by graded increasing nematode qualities. The current findings preliminarily revealed the gene expression pattern on the P. massoniana in response to B. xylophilus infestation.
-
Key words:
- Pinus massoniana /
- Bursaphelenchus xylophilus /
- gene expression /
- disease response
-
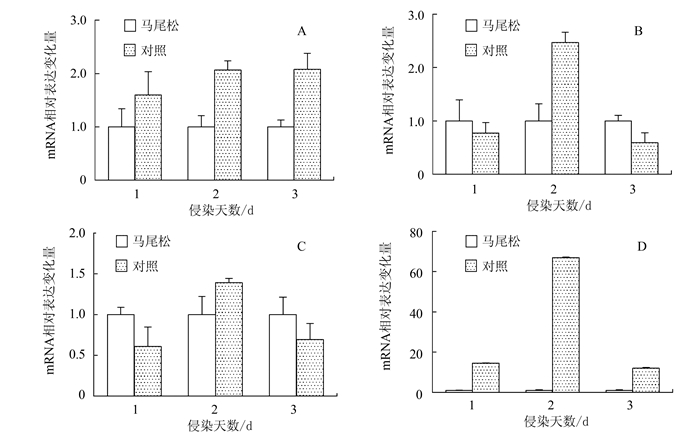
图 2 松材线虫侵染不同天数下的马尾松较其对照样本中的病原识别和抗逆调节基因的表达差异
注:A为CC-NBS-LRR抗性蛋白基因;B为late embryogenesis基因;C为class Ⅶ chitinase基因;D为class Ⅵ chitinase基因。
Figure 2. The differentially expressed genes relative to pathogen recognition and resistance regulation from the stems of P. massoniana infected by B. xylophilus in comparison with the corresponding control groups
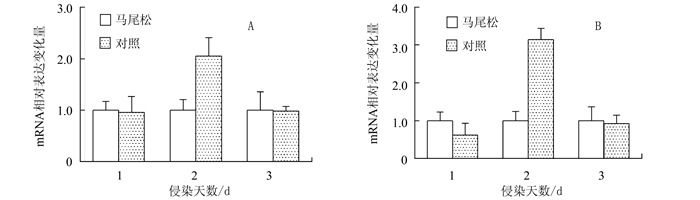
图 4 松材线虫侵染不同天数下的马尾松较其对照样本中的细胞色素P450基因的表达差异
注:A为cytochrome P450基因;B为cytochrome P450 mono-oxygenase superfamily基因;C为cytochrome P450 mono-oxygenase CYP7368基因。
Figure 4. The differentially expressed cytochrome P450 relevant genes from the stems of P. massoniana infected by B. xylophilus in comparison with the corresponding control groups
图 5 松材线虫侵染不同天数下的马尾松较其对照样本中的生长素响应基因的表达差异
注:A为auxin-responsive family protein基因;B为dormancy/auxin associated-like protein基因。
Figure 5. The differentially expressed genes involved in auxin-response from the stems of P. massoniana infected by B. xylophilus in comparison with the corresponding control groups
表 1 待测基因及其荧光定量PCR引物序列
Table 1. The candidate genes and their primers sequence using for qPCR
待测基因 上游引物序列 下游引物序列 dihydroflavonol-4-reductase 5′-GCGATGCCGCTATTGT-3′ 5′-TGAGGGCTTGCGAGAA-3′ CC-NBS-LRR resistance-like protein 5′-CCATAAGGATGTGAGAGCCA-3′ 5′-AAAGGTAGAAGTGCGAGAGG-3′ late embryogenesis abundant-like protein 5′-TGCCAAGTGGGACGATG-3′ 5′-TGGACGGTGACGCTGTTT-3′ class Ⅶ chitinase 5′-AGTTGAACCCTGCTCTGC-3′ 5′-GTGGAAACACCGACGAAG-3′ flavonoid 3-hydroxylase 5′-CAACGGCGTGAATGCTAAG-3′ 5′-AGAAGGAAGGCGTGGAGT-3′ class Ⅳ chitinase 5′-CGAGGGCAAGGGATTCTA-3′ 5′-ATTCCTGGCTGTTGATGGC-3′ auxin-responsive family protein 5′-ACGCTGCTGCTGTTCTCC-3′ 5′-CCAGGCTGTGGCATCTTC-3′ dormancy/auxin associated-like protein 5′-TTCAGGTCCCTTCTTTGC-3′ 5′-CTTTCGGTTTGTTTGCC-3′ cytochrome P450 mono-oxygenase superfamily 5′-AATCCGTCGTAGGCAACA-3′ 5′-GCCCGCCACATAGAAAT-3′ cytochrome P450 5′-GTCGGAAACCTCCACCAAC-3′ 5′-TAGGGACTGAGCCCAAGC-3′ cytochrome P450 monooxygenase CYP736B 5′-GTGGTCTTTGCTCCGTTAGGG-3′ 5′-AGCGTTGGCGTCATTCTGC-3′ actin 5′-CCTTGGCAATCCACATC-3′ 5′-TCACCACTACGGCAGAAC-3′ -
[1] ZHAO B G. Pine wilt disease in China[M]. Pine wilt disease Springer, 2008: 18-25. [2] XU L, LIU Z Y, ZHANG K, et al. Characterization of the Pinus massoniana transcriptional response to Bursaphelenchus xylophilus infection using suppression subtractive hybridization[J]. International Journal of Molecular Sciences, 2013, 14(6): 11356-11375. doi: 10.3390/ijms140611356 [3] ZHENG H Y, XU M, XU F Y, et al. A comparative proteomics analysis of Pinus massoniana inoculated with Bursaphelenchus xylophilus[J]. Pakistan Journal of Botany, 2015, 47(4): 1271-1280. [4] XIE W, HUANG A, LI H, et al. Identification and comparative analysis of microRNAs in Pinus massoniana infected by Bursaphelenchus xylophilus[J]. Plant Growth Regulation, 2016, doi: 10.1007/s10725-016-0221-8. [5] LⅣAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408. doi: 10.1006/meth.2001.1262 [6] HIRAO T, FUKATSU E, WATANABE A. Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization[J]. BMC Plant Biology, 2012, 12(1): 13. doi: 10.1186/1471-2229-12-13 [7] SANTOS C S, PINHEIRO M, SILVA A I, et al. Searching for resistance genes to Bursaphelenchus xylophilus using high throughput screening[J]. BMC Genomics, 2012, 13(1): 599. doi: 10.1186/1471-2164-13-599 [8] DANGL J L, JONES J D. Plant pathogens and integrated defence responses to infection[J]. Nature, 2001, 411(6839): 826-833. doi: 10.1038/35081161 [9] TAN S, WU S. Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon[J]. Comparative and Functional Genomics, 2012:ID 418208.doi: 10.1155/2012/418208. [10] ELLIS J, DODDS P, PRYOR T. Structure, function and evolution of plant disease resistance genes[J]. Current Opinion in Plant Biology, 2000, 3(4): 278-284. doi: 10.1016/S1369-5266(00)00080-7 [11] JONES J D, DANGL J L. The plant immune system[J]. Nature, 2006, 444(7117): 323-329. doi: 10.1038/nature05286 [12] HANIN M, BRINI F, EBEL C, et al. Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms[J]. Plant Signaling & Behavior, 2011, 6(10): 1503-1509. [13] YAMAMOTO T, IKETANI H, IEKI H, et al. Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens[J]. Plant Cell Reports, 2000, 19(7): 639-646. doi: 10.1007/s002999900174 [14] NEUHAUS J M. Plant chitinases (pr-3, pr-4, pr-8, pr-11)[M]. Pathogenesis-related proteins in plants CRC Press, 1999:77-105. [15] PETIT P, GRANIER T, D'ESTAINTOT B L, et al. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis[J]. Journal of Molecular Biology, 2007, 368(5): 1345-1357. doi: 10.1016/j.jmb.2007.02.088 [16] CHAPPLE C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases[J]. Annual Review of Plant Biology, 1998, 49(1): 311-343. doi: 10.1146/annurev.arplant.49.1.311 [17] RENAULT H, BASSARD J E, HAMBERGER B, et al. Cytochrome P450-mediated metabolic engineering: current progress and future challenges[J]. Current Opinion in Plant Biology, 2014, 19(19C): 27-34. -








 下载:
下载: