Genetic Comparisons Between Lactuca pulchella (Pursh) DC and Its Closely Related Lettuce Species
-
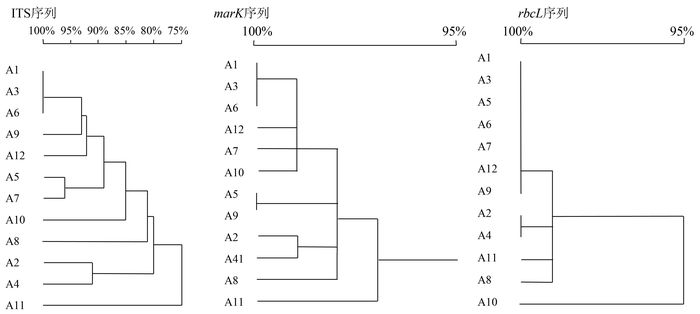
摘要: 利用PCR方法对野莴苣及其11种近似种DNA进行扩增,获得ITS、matK 和rbcL 序列,分析遗传相似系数和聚类分析图谱,探讨野莴苣及其近似种的亲缘关系。通过野莴苣及其近似种ITS序列分析发现:野莴苣和生菜、莴苣、毒莴苣的相似性分别为93.6%、93.1%、92.6%,野莴苣与乳苣的相似性仅为87.8%;matK 序列研究发现:野莴苣和生菜、莴苣、毒莴苣的相似性分别为99.2%、99.0%、99.1%,而野莴苣与乳苣的相似性仅为98.3%,结果表明,野莴苣与莴苣属的生菜、莴苣、毒莴苣的亲缘关系更近,与乳苣属的乳苣亲缘关系次之。通过对比rbcL 序列发现:野莴苣和生菜相似度达100%,与莴苣、翅果菊、毒莴苣、刺毛莴苣、宿根莴苣的相似度均达99.7%,而野莴苣与乳苣的相似度却为99.4%,将野莴苣归为莴苣属更加合理。Abstract: The genetic similarity coefficients and cluster analysis maps on Lactuca pulchella (Lp) and its 11 closely related species were obtained by using PCR to amplify ITS, matK and rbcL gene from the total DNA fragments of the lettuce varieties. Based on the ITS sequences, a similarity between Lp and L. sativa var. ramose was found to be 93.6%; between Lp and L. sativa , 93.1%; between Lp and L. serriola , 92.6%;and, between Lp and Mulgedium tataricum , the lowest at 87.8%. According to the matK sequences, a similarity between Lp and L. sativa var. ramose was shown to be 99.2%;between Lp and L. sativa , 99.0%, ; between Lp and L. serriol , 99.1%; and, between Lp and M.tataricum , 98.3%. It appeared that Lp was related closer to the Lactuca species than Mulgedium . By comparing the rbcL , Lp and L. sativa L. var. ramosa had a perfect match with a 100% similarity; whereas, Lp and L. sativa , L. indica , L.serriola , L. hirsute or L. perennis , at 99.7%;and, Lp and M. tataricum , at 99.4%. Consequently, it was concluded that Lp belonged to the genus Lactuca .
-
Key words:
- Lactuca pulchella /
- closely related species /
- genetic relationship
-
表 1 供试材料名称
Table 1. Test materials
试验编号 名称 拉丁名 A1 生菜 Lactuca sativa L. var. ramosa Hort. A2 苣荬菜 Sonchus arvensis L. A3 莴苣 L.sativa L. A4 苦苣菜 S. oleraceus L. A5 翅果菊 L. indica L. A6 毒莴苣 L. serriola L. A7 刺毛莴苣 L. hirsuta Muhl. ex Nutt. A8 黄鹌菜 Youngia japonica (L.)DC. A9 乳苣 Mulgedium tataricum (L.) DC. A10 宿根莴苣 L. perennis L. A11 雀苣 L. viminea L. A12 野莴苣 L.pulchella (Pursh) DC. 表 2 PCR所用引物序列
Table 2. Primers used for PCR
引物名称 引物序列(5'-3') 目标片断
/bpITS-F GGAAGTAAAAGTCGTAACAAG 722 ITS-R TCCTCCGCTTATTGATATGC matK -F ATACCTTACCCAGCCCATCT 580 matK -R AGCATTTGACTGCGTACCAT rbcL -F ATGTCACCACAAACAGAAAC 718 rbcL -R TCGCATGTACCTGCAGTAGC 表 3 PCR扩增反应体系
Table 3. PCR amplification reaction system
成分 体积/μ L 2×Taq PCR MasterMix 25 引物-F 2 引物-R 2 ddH2O 18 DNA template 3 总体积 50 表 4 野莴苣及其近似种ITS序列的遗传相似系数
Table 4. Genetic similarity coefficients of L. pulchella and its closely related species based on ITS sequences
(单位/%) 样品
编号A1 A2 A3 A4 A5 A6 A7 A8 A9 A10 A11 A12 A1 100 A2 81.0 100 A3 100 82.5 100 A4 80.3 91.2 81.9 100 A5 91.2 81.4 91.8 81.6 100 A6 100 82.8 100 82.1 91.9 100 A7 88.7 77.2 88.7 78.0 96.3 88.7 100 A8 82.6 80.6 83.9 80.4 82.1 84.1 77.2 100 A9 93.0 81.7 93.5 80.8 91.5 93.6 87.8 83.5 100 A10 85.7 78.4 85.4 77.1 87.0 85.6 86.1 78.0 85.0 100 A11 74.6 74.8 75.7 73.9 75.9 75.8 71.1 74.6 76.7 74.5 100 A12 93.6 78.4 93.1 77.3 87.3 92.6 84.6 80.4 87.8 82.5 74.2 100 注:A1为生菜;A2为苣荬菜;A3为莴苣;A4为苦苣菜;A5为翅果菊;A6为毒莴苣;A7为刺毛莴苣;A8为黄鹌菜;A9为乳苣;A10为宿根莴苣;A11为雀苣;A12为野莴苣。下表同。 表 5 野莴苣及其近似种matK 序列的遗传相似系数
Table 5. Genetic similarity coefficients of L. pulchella and its closely related species based on matK sequences
(单位/%) 样品
编号A1 A2 A3 A4 A5 A6 A7 A8 A9 A10 A11 A12 A1 100 A2 97.9 100 A3 99.8 98.0 100 A4 97.7 99.2 97.7 100 A5 98.1 96.7 98.6 96.9 100 A6 99.8 98.0 100 97.7 98.6 100 A7 98.6 97.5 99.0 97.1 97.9 98.9 100 A8 98.1 98.2 98.2 97.9 97.3 98.2 97.3 100 A9 98.1 96.7 98.6 96.9 100 98.6 97.9 97.3 100 A10 98.8 97.3 98.8 97.1 97.7 98.9 98.1 97.8 97.7 100 A11 96.7 97.3 97.4 96.7 96.7 97.3 97.0 97.6 96.7 96.5 100 A12 99.2 97.5 99.0 97.5 98.2 99.1 98.4 97.8 98.3 98.1 96.7 100 表 6 野莴苣及其近似种rbcL 序列的遗传相似系数
Table 6. Genetic similarity coefficients of L. pulchella and its closely related species based on rbcL sequences
(单位/%) 样品
编号A1 A2 A3 A4 A5 A6 A7 A8 A9 A10 A11 A12 A1 100 A2 99.1 100 A3 100 99.0 100 A4 99.0 100 99.1 100 A5 100 99.0 100 99.1 100 A6 100 99.0 100 99.1 100 100 A7 100 99.0 100 99.1 100 100 100 A8 98.9 99.2 99.0 99.2 99.0 99.0 99.0 100 A9 99.8 98.7 99.7 98.7 99.7 99.7 99.7 98.7 100 A10 100 93.0 93.6 99.1 93.9 94.0 93.6 93.5 93.7 100 A11 99.2 99.2 99.2 99.5 99.2 99.2 99.2 99.3 98.9 92.0 100 A12 100 98.8 99.7 99.1 99.7 99.7 99.7 98.6 99.4 99.7 98.9 100 -
[1] 中华人民共和国进境植物检疫性有害生物名录(更新至2017年6月, 441种)[OL]. [2017-06-05]. http://www.aqsiq.gov.cn/xxgk_13386/zvfg/gfxwj/dzwjy/201706/t20170614_490858.htm [2] Flora of North America Editorial Committee. Flora of North America, North of Mexico[M]. New York:Oxford University Press, 2006, 19-21:259. [3] Flora of Missouri Editorial Committee.Flora of Missouri[OL].[2017-06-05].http://www.efloras.org/florataxon.aspx?flora_id=11&taxon_id=200024120. [4] United States Department of Agriculture. Natural Resources Conservation Service.[OL].[2017-06-05]. http://plants.usda.gov/core/profile?symbol=LATAP. [5] 中国科学院中国植物志编辑委员会.中国植物志[M].北京:科学出版社, 1997, 80:75. [6] 韩亚光.新侵入辽宁地区的杂草——野莴苣[J].沈阳农业大学学报, 1995, 26(1):77-79. http://www.cqvip.com/qk/93761X/199501/index.shtml [7] 梁照文, 童明龙, 高振峰, 等.毒莴苣及其近似种的形态比较[J].植物检疫, 2014, 28(4):36-37. http://www.cqvip.com/QK/93407X/201404/661724391.html [8] Flora of China Editorial Committee.Flora of China (Asteraceae)[M]. Beijing & St. Louis:Science Press & Missouri Botanical Garden Press, 2011, 20-21:234, 237, 238. [9] KRESS W J, WURDACK K J, ZIMMER E A, et al. Use of DNA barcodes to identify flowering plants.[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(23):8369-8374. http://www.doc88.com/p-086650407796.html [10] TABERLET P, COISSAC E, POMPANON F, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding[J]. Nucleic Acids Research, 2007, 35(3):e14. doi: 10.1093/nar/gkl938 [11] KRESS W J, ERICKSON D L. A two-locus global DNA barcode for land plants:the coding rbcL gene complements the non-coding trnH-psbA spacer region[J]. Plos One, 2007, 2(6):e508. doi: 10.1371/journal.pone.0000508 [12] FAZEKAS A J, BRUGESS K S, KESANAKURTI P R, et al. Multiple Multilocus DNA Barcodes from the Plastid Genome Discriminate Plant Species Equally Well[J]. Plos One, 2007, 3(7):e2802. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.275.1938 [13] LAHAYE R, VAN D B M, BOGARIN D, et al. DNA barcoding the floras of biodiversity hotspots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(8):2923-2928. doi: 10.1073/pnas.0709936105 [14] 伏建国, 杨晓军, 钱路, 等.植物DNA条形码技术在出入境检验检疫领域的应用[J].植物检疫, 2012, 26(2):64-69. http://www.cqvip.com/qk/93407X/201202 [15] 付涛, 王志龙, 钱萍仙, 等.高等植物DNA条形码最新研究进展及其应用[J].核农学报, 2016, 30(5):887-896. doi: 10.11869/j.issn.100-8551.2016.05.0887 [16] FU Y M, JIANG W M, FU C X. Identification of species within Tetrastigma (Miq.) Planch.(Vitaceae) based on DNA barcoding techniques[J].植物分类学报, 2011, 49(3):237-245. http://www.cnki.com.cn/Article/CJFDTOTAL-ZWFX201103010.htm [17] LI D Z, GAO L M, LI H T, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants.[J]. Proceedings of the National Academy of Sciences, 2011, 108(49):19641-19646. doi: 10.1073/pnas.1104551108 [18] 高连明, 刘杰, 蔡杰, 等.关于植物DNA条形码研究技术规范[J].植物分类与资源学报, 2012, 34(6):592-60. http://ir.kib.ac.cn/handle/151853/16324 -







 下载:
下载:

