NPR1 Cloning and Expression during Development of Induced Systemic Resistance in Oncidium hybridum
-
摘要:
目的 探明文心兰NPR1基因在诱导抗性过程中的生理作用。 方法 利用RACE技术克隆文心兰NPR1基因全长,并进行相关生物信息学分析和遗传进化树的构建;以印度梨形孢共培养、水杨酸(SA)和茉莉酸甲酯(MeJA)处理文心兰,分析菊欧文氏杆菌导致的软腐病抗性影响及NPR1基因的表达特性变化。 结果 克隆得到的2条基因序列长度分别为1 804 bp和1 902 bp,编码555、556个氨基酸,均为NPR1类型,分别命名为OgNPR1-1和OgNPR1-2;文心兰两个NPR1基因在未接菌的情况下均呈现低表达量,在接种共生菌印度梨形孢后轻微上调表达,在接种菊欧文氏杆菌后显著上调表达,而在印度梨形孢共生菌的存在下,接种菊欧文氏杆菌使两个NPR1基因表达更为活跃;同时,相比印度梨形孢共生菌处理,两个NPR1基因均在外源SA、MeJA处理下接种菊欧文氏杆菌表现为更显著上调表达;此外,共生印度梨形孢可显著提高文心兰对菊欧文氏杆菌导致软腐病的抗性,限制软腐病病症在非接病叶片的扩展。 结论 在共生印度梨形孢的文心兰植株中,印度梨形孢显著提高了文心兰NPR1基因响应菊欧文氏杆菌的表达水平,虽然上调水平不及SA和MeJA激素处理的水平,却使文心兰植株表现出更强的对抗软腐病的能力。 Abstract:Objective Physiological role of NPR1 gene plays in the development of induced systemic resistance (ISR) in Oncidium hybridum was studied. Method NPR1 of the orchid was cloned using RACE technique, and bioinformatics and phylogenenetic analyses on the gene conducted. After co-cultured with Piriformospora indica or treated with salicylic acid (SA) or methyl jasmonate (MeJA) to induce ISR, the orchids were analyzed for the treatment effect against soft rot disease caused by Erwinia chrysanthemi. And, expressions of NPR1 during the treatments were investigated. Result Two full-length cDNA sequences were cloned. They were of a same type, one named OgNPR1-1 with a length of 1 804 bp encoding 555 amino acids, and the other, OgNPR1-2 1 902 bp encoding 556 amino acids. Their expressions were slightly upregulated by the inoculation of symbiotic P. indica. However, as both P. indica and E. chrysanthemi were present in the orchids, the expressions were significantly upregulated. It indicated that, in the presence of P. indica, the orchid plants became significantly more resistant to E. chrysanthemi inhibiting spread of the soft rot disease. On the other hand, the symbiosis of P. indica did not upregulated the expressions of the two genes more than the treatment by exogenous SA or MeJA on the orchids. Conclusion The expression of NPR1 in the orchids inoculated with the symbiotic P. indica was significantly upregulated in the presence of E. chrysanthemi. Although the increased expression by P. indica was lower than what induced by exogenous SA or MeJA hormone treatment, nonetheless, the orchids benefitted from the symbiosis and became more resistance to the soft rot disease. -
Key words:
- Oncidium hybridum /
- NPR1 /
- induced systemic resistance /
- Piriformospora indica /
- Erwinia chrysanthemi
-
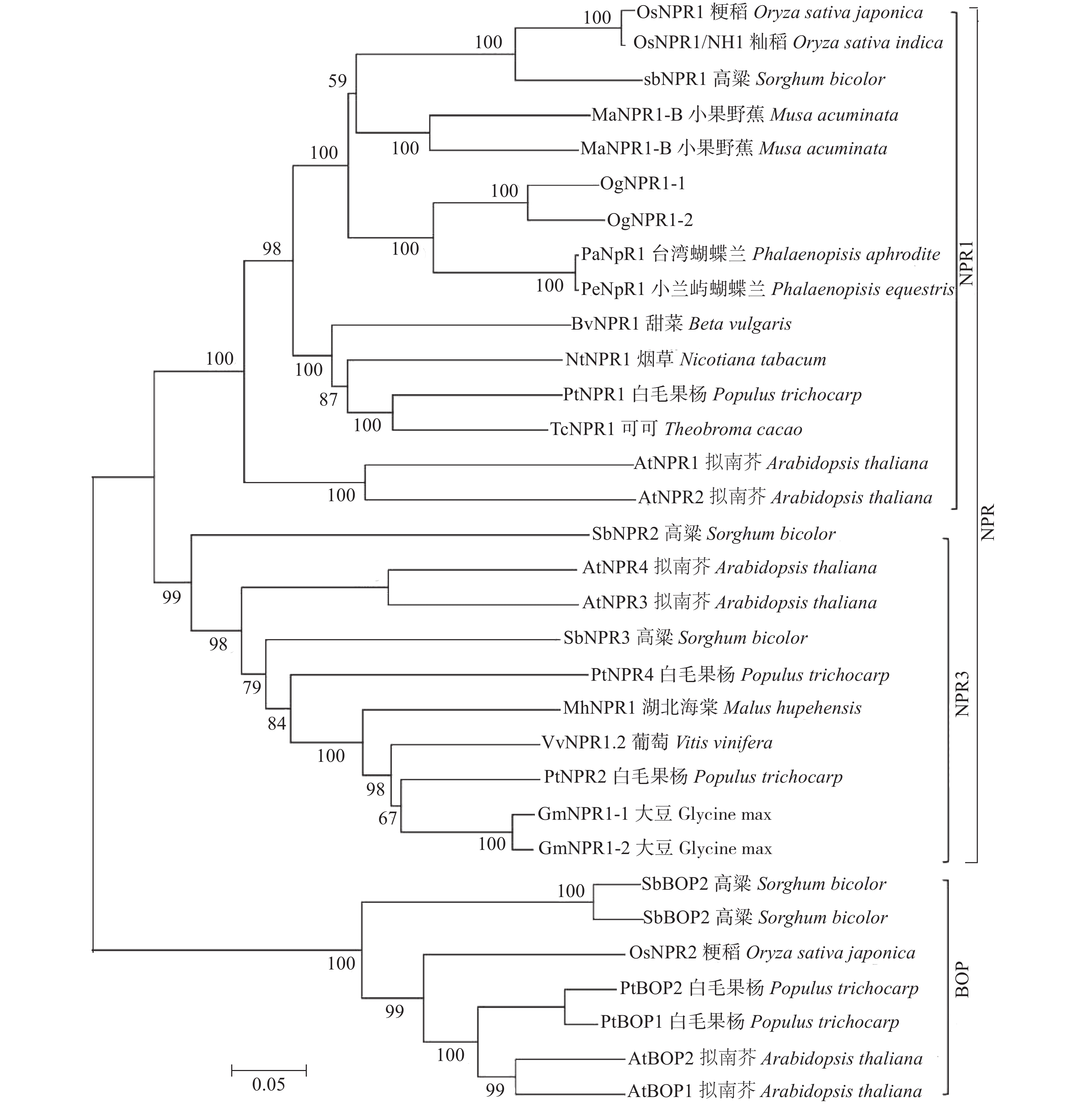
图 1 文心兰与其他物种的NPR1蛋白序列多重比对结果
注:Pa,百花蝴蝶兰;Pe,小兰屿蝴蝶兰;At,拟南芥;方框分别为NPR1氨基酸序列 Cys82 和Cys216位点。
Figure 1. Multiple sequence alignment of NPR1 from O. hybridum and other species
Note: Pa, Phalaenopsis amabilis; Pe, Phalaenopsis equestris; At, Arabidopsis thaliana; the characters with red frame are the Cys82 and Cys216 site of NPR1 protein sequences.
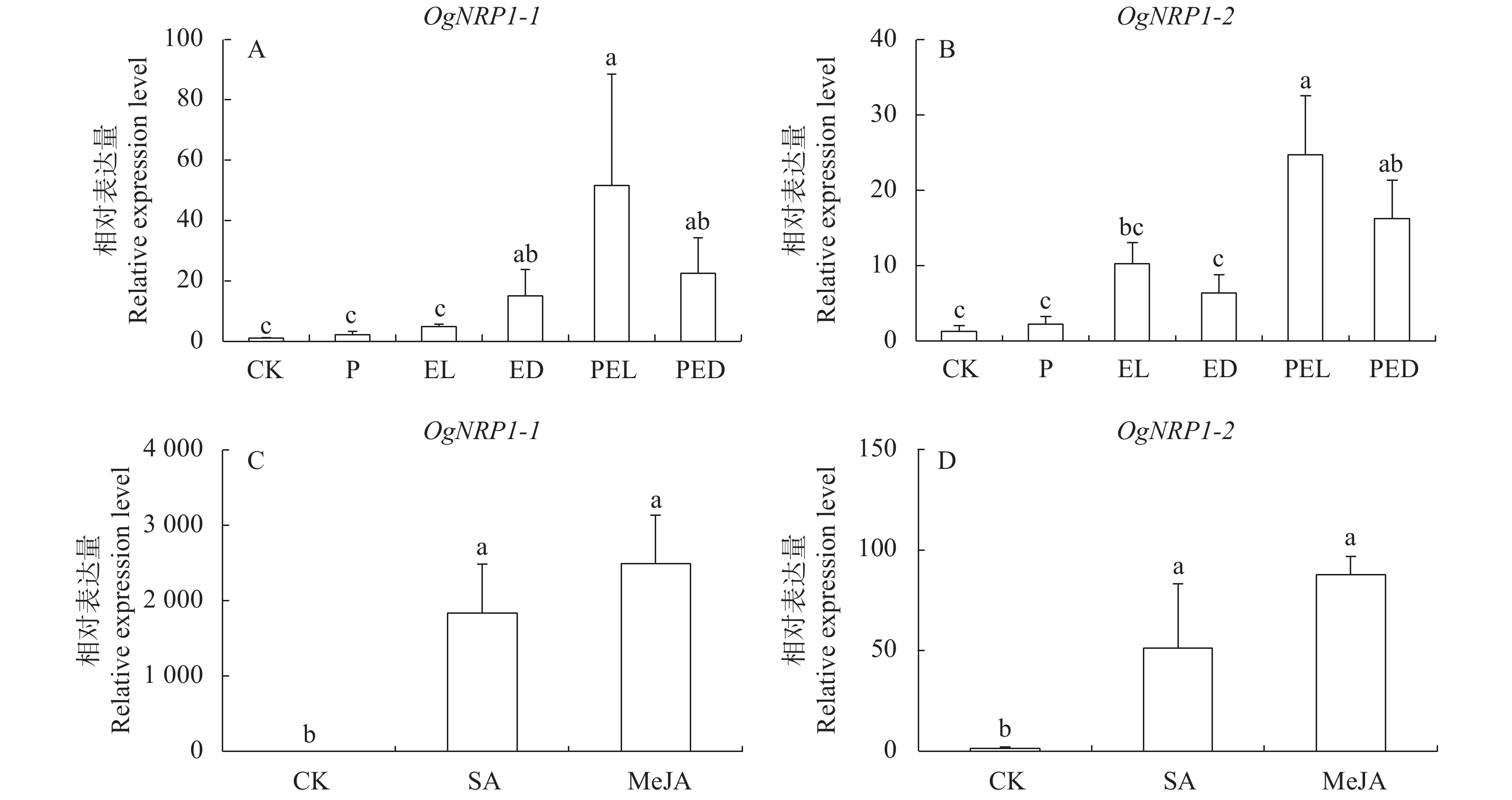
图 3 文心兰NPR1基因在ISR及外源激素处理下的表达模式
注:A、C,OgNPR1-1基因在不同处理条件下的相对表达量;B、D,OgNPR1-2基因在不同处理条件下的相对表达量;CK,对照组;P,印度梨形孢共培养处理文心兰叶片;EL,对照组文心兰接种病原菌24 h后接种叶片;ED,对照组文心兰接种病原菌24 h后邻近接种叶片;PEL,印度梨形孢共培养处理文心兰接种病原菌24 h后叶片;PED,印度梨形孢共培养处理文心兰接种病原菌24 h后邻近接种叶片;SA,喷施外源水杨酸24 h后处理叶片;MeJA,喷施外源茉莉酸酯24 h后处理叶片;不同字母代表不同处理间在P<0.05水平差异显著。
Figure 3. Expression patterns of NPR1 during ISR and exogenous hormone treatments on orchid plant
Note: A and C, relative expressions of OgNPR1-1 after different treatments; B and D, relative expressions of OgNPR1-2 after different treatments; CK, control; P, leaves of orchid plants co-cultured with P. indica; EL, leaves from control 24 h after inoculation with E. chrysanthemi; ED, adjacent leaves from control 24 h after inoculation with E. chrysanthemi; PEL, leaves from orchids co-cultured with P. indica 24 h after inoculation with E. chrysanthemi; PED, adjacent leaves from orchids co-cultured with P. indica 24 h after inoculation with E. chrysanthemi; SA, leaves treated with exogenous SA for 24 h; MeJA, leaves treated with exogenous MeJA for 24 h; different letters indicate significant differences (P<0.05).
图 4 印度梨形孢及外源激素处理后文心兰对软腐病抗性影响
注:CK,对照;MeJA,茉莉酸酯;SA,水杨酸;P,印度梨形孢共培养。
Figure 4. Effects of P. indica and exogenous hormone treatments on resistance of O. hybridum against soft rot disease
Note: CK, Control group; MeJA, Oncidium plants treated with exogenous MeJA; SA, Oncidium plants treated with exogenous SA; P, Oncidium plants co-cultured with P. indica.
表 1 引物列表
Table 1. List of primers
引物 Primer 序列 Sequences(5'-3') RACE扩增 RACE amplification NPR1_F1 CCGATGTCAACCATAAGAACCA NPR1_F2 GCTCTTTCCAGAACAGTTGAAC NPR1_R1 GTGGATGATGATGATGACAAGCC NPR1_R2 TTCAGAACCTCCGAGCATCG 3' RACE Outer Primer GCGAGCACAGAATTAATACGACT 3' RACE Inner Primer CGCGGATCCGAATTAATACGACTCACTATAGG 5' RACE Outer Prime GCTGATGGCGATGAATGAACACTG 5' RACE Inner Primer CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG 开放阅读框 ORF Onc NPR1-1 1F ATGATCGCCACCACCTTCTCC Onc NPR1-2 1F ATGACCGCCACCGCCCTCTCA Onc NPR1-1 1665R CTAAAATAAATTACTTGCCTGAAC Onc NPR1-2 1668R CTAAAATAATTTATTTGCCTGAAC qPCR扩增 qPCR amplification Onc NPR1-1 15F TTCTCCGACTCCGACAACAC Onc NPR1-1 182R ACTCGATTGATCCGACGGT Onc NPR1-2 15F CCCTCTCAGACTCCGACAACTT Onc NPR1-2 182R AAACTCGAT TAAGCCGACG AC actin F291 GGAGAAACTTGCTTATGTGGC actin R399 CGTAATGACTTGACCATCCG 表 2 文心兰NPR1氨基酸序列Blastp结果
Table 2. Blastp results of Oncdium NPR1 amino acid sequences
序列 Sequences 物种 Species 注册号 Accession 一致性 Consistency/% E值 E value OgNPR1-1 Phalaenopsis aphrodite subsp. formosana AEP68016.1 81.12 0 OgNPR1-2 Phalaenopsis equestris XP_020587799.1 79.59 0 -
[1] KOGEL K H, LANGEN G. Induced disease resistance and gene expression in cereals [J]. Cellular Microbiology, 2005, 7(11): 1555−1564. [2] MOLITOR A, ZAJIC D, VOLL L M, et al. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica–mediated systemic induced resistance to powdery mildew [J]. Molecular Plant-Microbe Interactions, 2011, 24(12): 1427−1439. doi: 10.1094/MPMI-06-11-0177 [3] GLAESER S P, IMANI J, ALABID I, et al. Non-pathogenic Rhizobium radiobacter F4 deploys plant beneficial activity independent of its host Piriformospora indica [J]. The ISME Journal, 2016, 10(4): 871−884. doi: 10.1038/ismej.2015.163 [4] NASSIMI Z, TAHERI P. Endophytic fungus Piriformospora indica induced systemic resistance against rice sheath blight via affecting hydrogen peroxide and antioxidants [J]. Biocontrol Science and Technology, 2017, 27(2): 252−267. doi: 10.1080/09583157.2016.1277690 [5] HANEY C H, WIESMANN C L, SHAPIRO L R, et al. Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens [J]. Molecular Ecology, 2018, 27(8): 1833−1847. doi: 10.1111/mec.14400 [6] NIE P P, LI X, WANG S N, et al. Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET-and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis [J]. Frontiers in Plant Science, 2017, 8: 238−250. doi: 10.3389/fpls.2017.00238 [7] WALLER F, ACHATZ B, BALTRUSCHAT H, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield [J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(38): 13386−13391. doi: 10.1073/pnas.0504423102 [8] SCHÄFER P, PFIFFI S, VOLL L M, et al. Phytohormones in plant root-Piriformospora indica mutualism [J]. Plant Signaling & Behavior, 2009, 4(7): 669−671. [9] SCHÄFER P, PFIFFI S, VOLL L M, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica [J]. The Plant Journal, 2009, 59(3): 461−474. doi: 10.1111/j.1365-313X.2009.03887.x [10] SCHÄFER P, KOGEL K H. The sebacinoid fungus piriformospora indica: an orchid mycorrhiza which may increase host plant reproduction and fitness[M].The Mycota. Berlin, Heidelberg: Springer Berlin Heidelberg, : 99-112. [11] PEDROTTI L, MUELLER M J, WALLER F. Piriformospora indica root colonization triggers local and systemic root responses and inhibits secondary colonization of distal roots [J]. PLoS One, 2013, 8(7): e69352. doi: 10.1371/journal.pone.0069352 [12] LIN H F, XIONG J, ZHOU H M, et al. Growth promotion and disease resistance induced in Anthurium colonized by the beneficial root endophyte Piriformospora indica [J]. BMC Plant Biology, 2019, 19: 40−51. doi: 10.1186/s12870-019-1649-6 [13] GILL S S, GILL R, TRIVEDI D K, et al. Piriformospora indica: potential and significance in plant stress tolerance [J]. Front Microbiology, 2016, 7: 332. doi: 10.3389/fmicb.2016.00332 [14] JIANG H Y, ZHANG J L, YANG J W, et al. Transcript profiling and gene identification involved in the ethylene signal transduction pathways of creeping bentgrass (Agrostis stolonifera) during ISR response induced by butanediol [J]. Molecules, 2018, 23(3): 706−118. doi: 10.3390/molecules23030706 [15] MHAMDI A. NPR1 has everything under control [J]. Plant Physiology, 2019, 181(1): 6−7. doi: 10.1104/pp.19.00890 [16] CAO H, BOWLING S A, GORDON A S, et al. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance [J]. The Plant Cell, 1994, 6(11): 1583−1592. doi: 10.2307/3869945 [17] CAO H, GLAZEBROOK J, CLARKE J D, et al. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats [J]. Cell, 1997, 88(1): 57−63. doi: 10.1016/S0092-8674(00)81858-9 [18] BACKER R, NAIDOO S, VAN DEN BERG N. The nonexpressor of pathogenesis-related genes 1 (NPR1) and related family: mechanistic insights in plant disease resistance [J]. Frontiers in Plant Science, 2019, 10: 102−113. doi: 10.3389/fpls.2019.00102 [19] DING Y L, SUN T J, AO K, et al. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity [J]. Cell, 2018, 173(6): 1454−1467. doi: 10.1016/j.cell.2018.03.044 [20] KINKEMA M, FAN W H, DONG X N. Nuclear localization of NPR1 is required for activation of PR gene expression [J]. The Plant Cell, 2000, 12(12): 2339−2350. doi: 10.1105/tpc.12.12.2339 [21] ZHANG Y, FAN W, KINKEMA M, et al. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene [J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(11): 6523−6528. doi: 10.1073/pnas.96.11.6523 [22] ZHOU J M, TRIFA Y, SILVA H, et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1Gene required for induction by salicylic acid [J]. Molecular Plant-Microbe Interactions, 2000, 13(2): 191−202. doi: 10.1094/MPMI.2000.13.2.191 [23] FAN W H, DONG X N. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis [J]. The Plant Cell, 2002, 14(6): 1377−1389. doi: 10.1105/tpc.001628 [24] SKELLY M J, FURNISS J J, GREY H L, et al. Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1 [J]. Elife Science, 2019, 8(3): 146−157. [25] XUE D, GUO N, ZHANG X L, et al. Genome-wide analysis reveals the role of mediator complex in the soybean-Phytophthora sojae interaction [J]. International Journal of Molecular Sciences, 2019, 20(18): 4570−4582. doi: 10.3390/ijms20184570 [26] LAI Y S, RENNA L, YAREMA J, et al. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic Reticulum [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(22): 5203−5212. doi: 10.1073/pnas.1802254115 [27] PIETERSE C M J, VAN WEES S C M, VAN PELT J A, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis [J]. The Plant Cell, 1998, 10(9): 1571−1580. doi: 10.1105/tpc.10.9.1571 [28] GLAZEBROOK J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens [J]. Annual Review of Phytopathology, 2005, 43(1): 205−227. doi: 10.1146/annurev.phyto.43.040204.135923 [29] SPOEL S H, KOORNNEEF A, CLAESSENS S M C, et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol [J]. The Plant Cell, 2003, 15(3): 760−770. doi: 10.1105/tpc.009159 [30] POKOTYLO I, KRAVETS V, RUELLAND E. Salicylic acid binding proteins (SABPs): the hidden forefront of salicylic acid signalling [J]. International Journal of Molecular Sciences, 2019, 20(18): 4377. doi: 10.3390/ijms20184377 [31] RAYAPURAM C, BALDWIN I T. Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature [J]. The Plant Journal, 2007, 52(4): 700−715. doi: 10.1111/j.1365-313X.2007.03267.x [32] TANG J Y, WANG Y Q, YIN W C, et al. Mutation of a nucleotide-binding leucine-rich repeat immune receptor-type protein disrupts immunity to bacterial blight [J]. Plant Physiology, 2019, 181(3): 1295−1313. doi: 10.1104/pp.19.00686 [33] SU C C, LEU L S. Soft rot of Oncidium "Gower Ramsey" and Cymbidium sp. caused by Erwinia carotovora subsp. carotovora [J]. Plant Pathology Bulletin, 1992, 1(4): 190−195. [34] SAMSON R. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(4): 1415-1427. [35] 杨学, 叶炜, 李永清, 等. 南茜文心兰软腐病病原分离鉴定及致病性分析 [J]. 亚热带植物科学, 2017, 46(3):201−208. doi: 10.3969/j.issn.1009-7791.2017.03.001YANG X, YE W, LI Y Q, et al. Isolation, identification and pathogenicity analysis of soft rot pathogen from Oncidium ‘Gower Ramsey’ [J]. Subtropical Plant Science, 2017, 46(3): 201−208.(in Chinese) doi: 10.3969/j.issn.1009-7791.2017.03.001 [36] HUA MOD S, SENTHIL KUMAR R, SHYUR L F, et al. Metabolomic compounds identified in Piriformospora indica-colonized Chinese cabbage roots delineate symbiotic functions of the interaction [J]. Scientific Reports, 2017, 7(1): 9291−9303. doi: 10.1038/s41598-017-08715-2 [37] GUO H J, GLAESER S P, ALABID I, et al. The abundance of endofungal bacterium Rhizobium radiobacter (syn. Agrobacterium tumefaciens) increases in its fungal host piriformospora indica during the tripartite sebacinalean symbiosis with higher plants [J]. Frontiers in Microbiology, 2017, 8(3): 629−641. doi: 10.3389/fmicb.2017.00629 [38] YE W, SHEN C H, LIN Y L, et al. Growth promotion-related miRNAs in Oncidium orchid roots colonized by the endophytic fungus piriformospora indica [J]. PLoS One, 2014, 9(1): e84920. doi: 10.1371/journal.pone.0084920 [39] CHEN J C, LU H C, CHEN C E, et al. The NPR1 ortholog PhaNPR1 is required for the induction of PhaPR1 in Phalaenopsis aphrodite [J]. Botanical Studies, 2013, 54(1): 31−41. doi: 10.1186/1999-3110-54-31 [40] INNES R. The positives and negatives of NPR: a unifying model for salicylic acid signaling in plants [J]. Cell, 2018, 173(6): 1314−1315. doi: 10.1016/j.cell.2018.05.034 [41] 江金兰. 不同添加物对文心兰原球茎增殖及试管苗生长的影响 [J]. 亚热带植物科学, 2012, 41(4):8−14.JIANG J L. Study on the effects of several additives on the PLB proliferation and tube seedlings growth of Oncidium 'Gower Ramsey' [J]. Subtropical Plant Science, 2012, 41(4): 8−14.(in Chinese) [42] CAO H, LI X, DONG X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance [J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(11): 6531−6536. doi: 10.1073/pnas.95.11.6531 [43] HEPWORTH S R, ZHANG Y L, MCKIM S, et al. BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis [J]. The Plant Cell, 2005, 17(5): 1434−1448. doi: 10.1105/tpc.104.030536 [44] KHAN M, XU H S, HEPWORTH S R. BLADE-ON-PETIOLE genes: Setting boundaries in development and defense [J]. Plant Science, 2014, 215/216: 157−171. doi: 10.1016/j.plantsci.2013.10.019 [45] PALMER I A, CHEN H, CHEN J, et al. Novel salicylic acid analogs induce a potent defense response in Arabidopsis [J]. International Journal of Molecular Sciences, 2019, 20(13): 3356−3368. doi: 10.3390/ijms20133356 [46] SHU L J, LIAO J Y, LIN N C, et al. Identification of a strawberry NPR-like gene involved in negative regulation of the salicylic acid-mediated defense pathway [J]. PLoS One, 2018, 13(10): e0205790. doi: 10.1371/journal.pone.0205790 [47] MOU Z L, FAN W H, DONG X N. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes [J]. Cell, 2003, 113(7): 935−944. doi: 10.1016/S0092-8674(03)00429-X [48] ZHANG W, CORWIN J A, COPELAND D, et al. Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem [J]. The Plant Cell, 2017, 29(11): 2727−2752. doi: 10.1105/tpc.17.00348 [49] ENDAH R, BEYENE G, KIGGUNDU A, et al. Elicitor and Fusarium-induced expression of NPR1-like genes in banana [J]. Plant Physiology and Biochemistry, 2008, 46(11): 1007−1014. doi: 10.1016/j.plaphy.2008.06.007 [50] 任陪娣, 谢碧玉, 陈翠, 等. 香蕉NPR1基因家族的鉴定及在枯萎病菌胁迫下表达分析 [J]. 植物遗传资源学报, 2019, 20(6):1621−1629.REN P D, XIE B Y, CHEN C, et al. Genome-wide survey of the NPR1 gene family in banana(Musa spp.) and expression pattern analysis upon infection with foc TR4 [J]. Journal of Plant Genetic Resources, 2019, 20(6): 1621−1629.(in Chinese) [51] YUAN Y X, ZHONG S H, LI Q, et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility [J]. Plant Biotechnology Journal, 2007, 5(2): 313−324. doi: 10.1111/j.1467-7652.2007.00243.x -








 下载:
下载: