Transcriptome Analysis on Cryotherapy-treated Virus-free Plantlets of Red Bud Taro at Yanshan, Jiangxi Province
-
摘要:
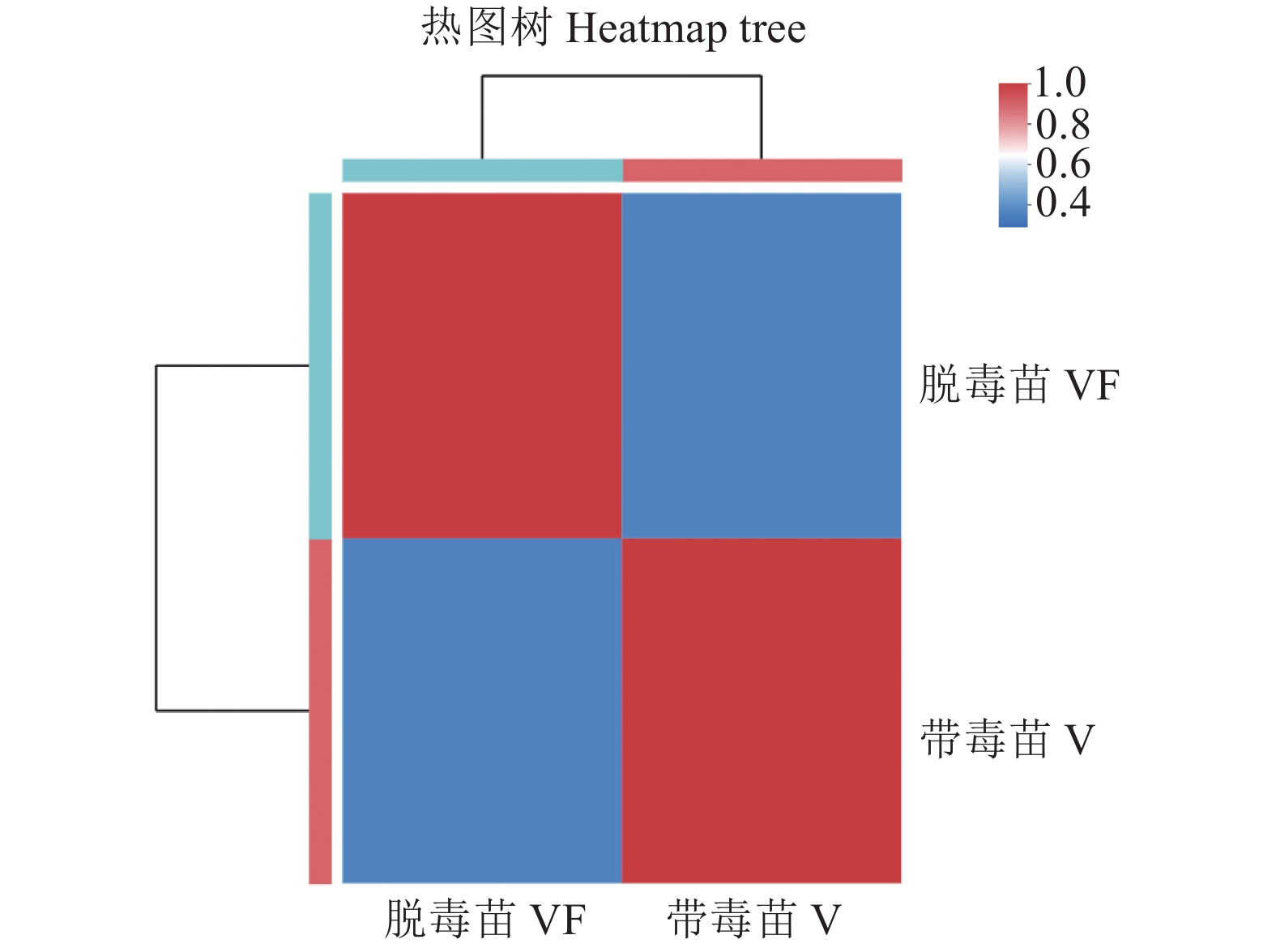
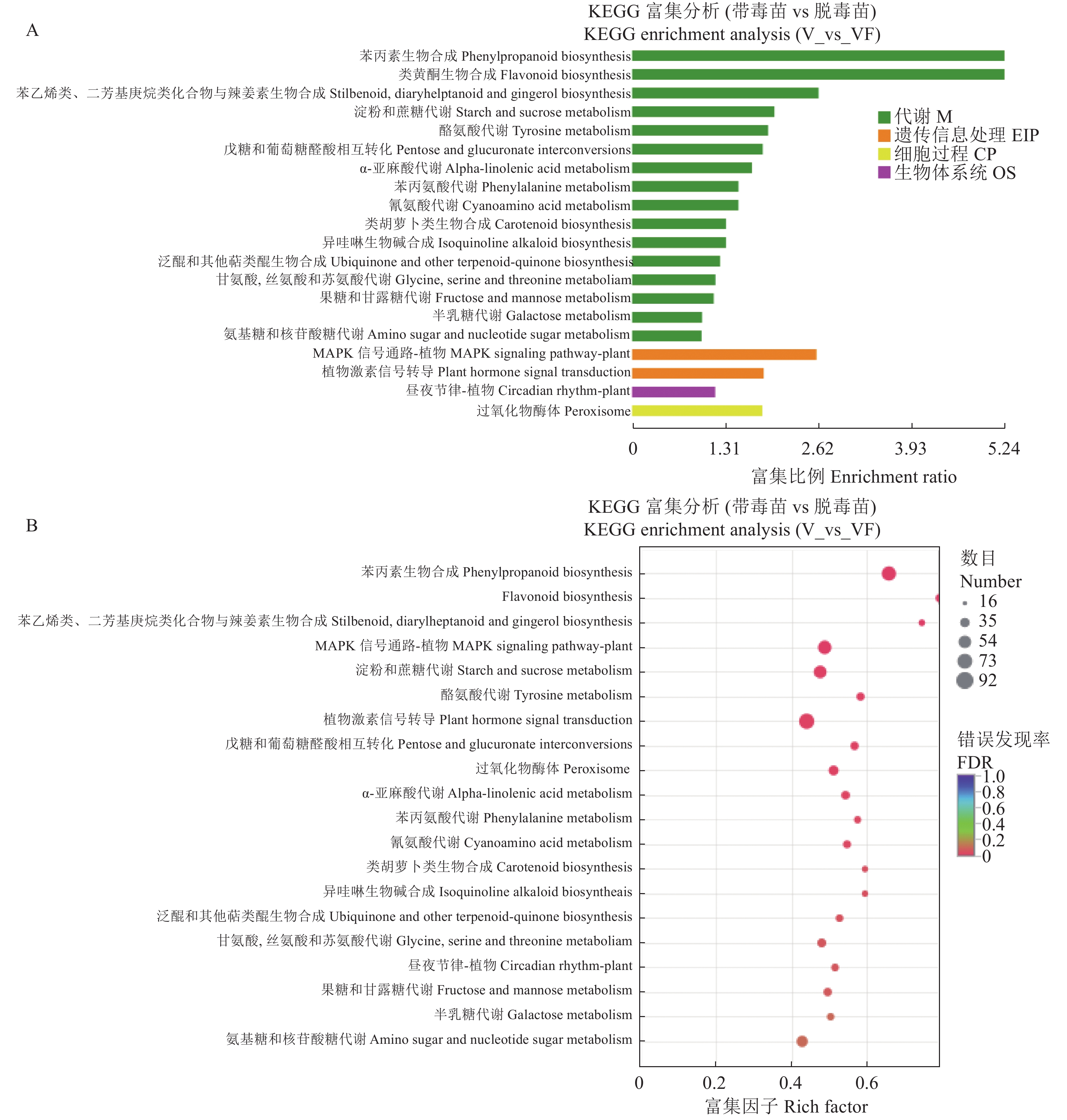
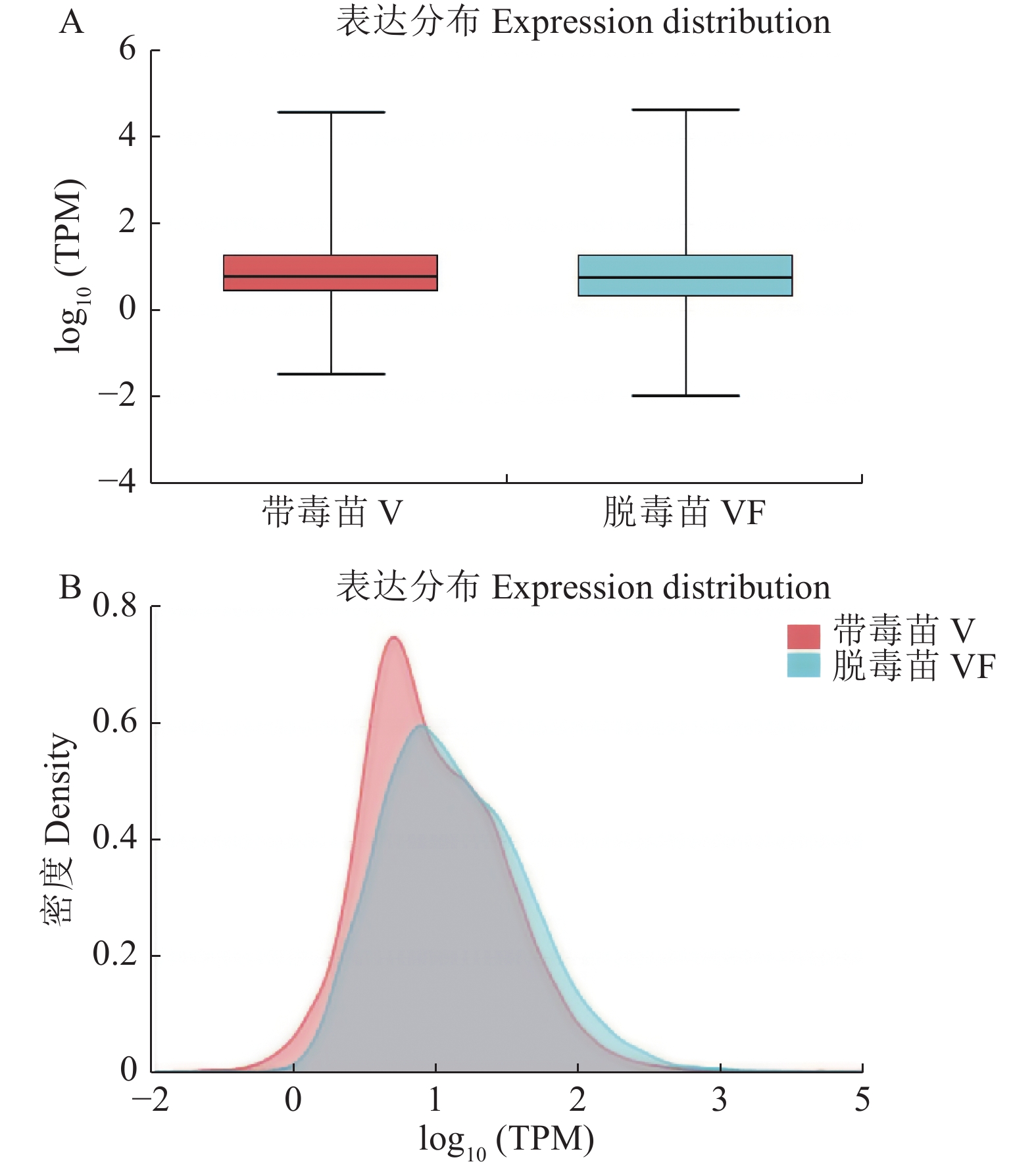
目的 探究江西铅山红芽芋对超低温疗法脱毒的转录组响应机制,为江西铅山红芽芋超低温疗法脱毒苗的规模化应用奠定理论依据。 方法 以江西铅山红芽芋超低温疗法脱毒苗(VF组)和江西铅山红芽芋超低温疗法带毒苗(V组)试验材料进行转录组比较分析。 结果 VF组Clean reads为42 406 188,GC含量为54.09%;V组Clean reads为46 818 060,GC含量为50.36%。VF组和V组表达量FPKM的对数值在0~2,表达量密度在0~0.8。VF组和V组表达的共有基因数为23 820,V组单独表达的基因数为10 298,VF组单独表达的基因数为4 477。VF组和V组表达量的相关系数为0.287,样本间相关性较差。VF组和V组共产生差异表达基因5 282 个,与V组比较,VF组上调基因3 011,下调基因2 271。GO 富集分析显示,差异基因主要注释到过氧化氢分解过程、过氧化氢代谢过程、一元羧酸生物合成过程、多糖分解代谢过程、金属离子输运、细胞外区、细胞壁、外部封装结构、膜的固有成分、质膜固有成分、核酸结合转录因子活性、序列特异性DNA结合转录因子活性、单加氧酶活性、铁离子结合、血红素结合等功能。KEGG 富集分析显示,差异基因主要注释到苯丙酸生物合成、类黄酮生物合成、二苯乙烯类和二芳基庚烷类以及姜辣素的生物合成、MAPK信号通路-植物、淀粉和蔗糖代谢、酪氨酸代谢、植物激素信号转导、戊糖和葡萄糖醛酸的相互转化、过氧化物酶体、α-亚麻酸代谢、苯丙氨酸代谢、氰胺酸代谢、类胡萝卜素生物合成、异喹啉生物碱的生物合成、泛醌和其他萜类醌的生物合成、甘氨酸、丝氨酸和苏氨酸代谢、植物昼夜节律、果糖和甘露糖代谢、半乳糖代谢、氨基糖和核苷酸糖代谢等途径。 结论 F-box家族蛋白、脱落酸受体PYR1、乙烯受体2、生长素响应因子1、BZIP转录因子家族蛋白、过氧化物酶、过氧化氢酶1、超氧化物歧化酶[Cu-Zn]、植物抗病反应蛋白、抗病蛋白、抗病蛋白RPS2、抗病蛋白RPS5、抗病蛋白RGA2、抗病反应蛋白、转座子TNT的逆转录病毒相关Pol多蛋白、逆转录病毒相关Pol多聚蛋白系1、类烟草病毒增殖蛋白3、烟草病毒增殖蛋白1、转座子RE1的逆转录病毒相关Pol多蛋白、蔗糖合成酶等基因是江西铅山红芽芋的超低温疗法脱毒的主要响应基因。 Abstract:Objective Mechanism of red bud taro in response to cryotherapy was studied by transcriptome analysis for extended application of the virus-free seedlings at Yanshan, Jiangxi. Method Transcriptomes of virus-free (VF) and virus-carrying (V) red bud taro plantlets that had been cryo-therapeutically treated were analyzed and compared. Result In the VF plantlets, the clean reads were 42 406 188 with a GC content of 54.09%; while, in the V plantlets, the clean reads were 46 818 060 with a GC content of 50.36%. For either group, the FPKM was between 0-2, and the expression density between 0-0.8. There were 23 820 genes commonly expressed in both VF and V, while 4 477 independently expressed in VF, and 10 298 in V. There was a low correlation between VF and V with a coefficient of 0.287, and were 5 282 differentially expressed genes (DEG) in them. Compared to V, VF had 3 011 genes were up-regulated and 2 271 down-regulated. The Go enrichment analysis showed that the differential genes were annotated mainly in the processes of hydrogen peroxide catabolism, hydrogen peroxide metabolism, monocarboxylic acid biosynthesis, and polysaccharide catabolism, metal ion transport, extracellular region, cell wall, external encapsulating structure, intrinsic components of membrane and plasma membrane as well as the activities of nucleic acid binding transcription factor, transcription factor, sequence-specific DNA binding, monooxygenase, iron ion binding, heme binding, etc. The KEGG enrichment analysis indicated that the differential genes were annotated largely to the biosynthesis of phenylpropanoid, flavonoid, stilbenoid, diarylheptanoid, gingerol, carotenoid, isoquinoline alkaloid, ubiquinone, and other terpenoid-quinones, the metabolisms of starch, sucrose, tyrosine, alpha-Linolenic acid, phenylalanine, cyanoamino acid, glycine, serine, threonine, fructose, mannose, galactose, amino sugar, and nucleotide sugar, and the pathways of MAPK signaling-plant, plant hormone signal transduction, pentose and glucuronate interconversions, peroxisome, circadian rhythm-plant, and others, Conclusion The genes of F-box family protein, abscisic acid receptor PYR1, ethylene receptor 2, auxin response factor 1, BZIP transcription factor family protein, peroxidase, catalase 1, superoxide dismutase [Cu-Zn], plant disease resistance response protein, disease resistance protein, disease resistance protein RPS2, disease resistance protein RPS5, disease resistance protein RGA2, disease resistance response protein, retrovirus-related Pol polyprotein from transposon TNT, retrovirus-related Pol polyprotein LINE-1, tobamovirus multiplication protein 3-like, tobamovirus multiplication protein 1, retrovirus-related Pol polyprotein from transposon RE1, sucrose synthase, etc. were the predominant genes in the virus-free red bud taro plantlets that responded to the cryotherapy. -
表 1 qRT-PCR引物序列
Table 1. Primers designed for qRT-PCR
基因
Gene方向
Direction引物序列
Primer sequence(5′-3′)大小
Size/bpTM值
TM value抗病反应蛋白 Disease resistance response protein F TTCGCGACGAAAGGTAAAACGA 105 55 R CACCCACAGGATGGACCAGAGG 抗病蛋白RGA2 Disease resistance protein RGA2 F CGGTTCAGAAGGGAGCCTATTGT 199 53.3 R AAACTCTTATACCTTTGGGGGGC 乙烯受体2 Ethylene receptor 2 F TCCCTTTTGACCTTGTTCTTTTG 137 52 R TACATTCTCCCGTGTACTTGCAG 过氧化氢酶1 Catalase 1 F CAGTGTCAAGACACGGTCGCATG 198 53.8 R CTAGTAACCGCTAGGGCCGTTGG 蔗糖合成酶 Sucrose synthase F GCCCCGTCTTATTCATACCCTCG 173 54.6 R CATGCTTTTCCCATTTTGTCCTCA 抗病蛋白RPS5 Disease resistance protein RPS5 F GTCCCAACTTATCCGTCATTTCT 168 51.4 R TAAAGGAGCTGCCACTCACAGTC 抗病蛋白 Disease resistance protein F TGTGTCTCCCTCCAGTGCTCA 145 53.6 R TTTCTCGCCCCCCCTGTTCA 类烟草病毒增殖蛋白3 Tobamovirus multiplication protein 3-like F GGTGTCTCGTTGTTTGCTGCTC 103 52.1 R GTTTCTTGCGTCGCCCTTTC 植物抗病反应蛋白 Plant disease resistance response protein F TGCCCTGAGCGTTCCCTAC 178 58.3 R ACCCGCCTCCACTTCTTCC F-box家族蛋白 F-box family protein F CGCCAAGAGGGAAATGAAACC 195 58.5 R CGGGAACATGGAGCAGTGGTA 超氧化物歧化酶[Cu-Zn] Superoxide dismutase [Cu-Zn] F CTGAGGCAACGATTGTGGATA 130 53.2 R CCCGTGCTAAGGCTAAGTTCAT 逆转录病毒相关Pol多聚蛋白系-1
Retrovirus-related Pol polyprotein line-1F CCCTTCTTTGTTAATGCCCACCA 156 49.9 R CGTCCTTAATGCACGGAAGCA 转座子RE1的逆转录病毒相关Pol多蛋白
Retrovirus-related Pol polyprotein from transposon RE1F GTCCAGATTGAAGTCGCTCCC 117 53.5 R CCTTGTACTGTGACAACAATGCC 转座子TNT逆转录病毒相关Pol多蛋白
Retrovirus-related Pol polyprotein from transposon TNTF GATAGACCATCAGGCACTTTAGG 151 48.5 R CTAAGTCATTCTTGGGACACTGTAA BZIP转录因子家族蛋白 BZIP transcription factor family protein F CATCGCCGAGCACTTGCACCGTCTC 95 55.6 R GCTTCTGGCATCTTTACCACTTTC 过氧化物酶 Peroxidase F CCAACTCCCAGGACTTCTTCTTCC 136 57.9 R CTGGCTGGCTAGTGGTGCTTGTT 脱落酸受体PYR1 Abscisic acid receptor PYR1 F CTCCATTTCGCCCTGATTCCATT 139 54.6 R TCAAACATCCCTTTCACCATTCCTAT 抗病蛋白RPS2 Disease resistance protein RPS2 F ATCCATTCCTTGCCAGATGAC 115 53.4 R ACCTTATTAACCGCAGCACCA 生长素反应因子1 Auxin response factor 1 F CTTTCACGCCCAATCGACCAC 169 57.6 R TCGGCTTCTTGCTTTCTGCTGTC 烟草病毒增殖蛋白1 Tobamovirus multiplication protein 1 F CTGTGGGAAGACTTCTTGCCTGAT 108 50.9 R TCGCTTTGGTATTATTGGACCTG 甘油醛-3-磷酸脱氢酶 GAPDH (内参 Reference gene) F ATCAAGCCCTCAACAATGCCAAA 179 54.8 R GCCAAGAAGGTCGTCATCTCAGC 表 2 V组和VF组测序数据统计
Table 2. Statistical sequencing data on V and VF plantlets
样本 Sample 带毒苗 V 脱毒苗 VF 原始序列
Raw reads47 418 842 43 025 444 原始总碱基数
Raw bases7 160 245 142 6 496 842 044 过滤后序列
Clean reads46 818 060 42 406 188 质控后总碱基数
Clean bases6 964 653 736 6 305 931 529 测序错误率
Error rate/%0.023 1 0.024 6 大于20的碱基数占总碱基的百分比
Q20/%98.83 98.26 大于30的碱基数占总碱基的百分比
Q30/%96.08 94.50 G、C碱基的总数量占总碱基数量的百分比
GC content/%50.36 54.09 表 3 测序数据与组装结果比对统计
Table 3. Comparison on sequencing data and assembly results
样本
Sample过滤后测序
数据的条数
Clean reads能比对到组装
转录本上的
Clean reads数
Mapped reads能定位到组装转
录本上的Clean
reads所占百分比
Mapped ratio/%脱毒苗VF 42 406 188 34 683 956 81.79 带毒苗V 46 818 060 38 036 872 81.24 表 4 VF组和V组表达量差异统计(前10)
Table 4. Top 10 statistical expression differences between VF and V plantlets
基因编号
Gene_ID差异表达倍数
FC(VF/V)差异表达倍数
以2为底的
对数值
log2FC
(VF/V)P值
P value矫正后 P值
P adjust显著性
Significance调节
Regulate带毒苗
表达量
Expression
in V脱毒苗
表达量
Expression
in VFNR描述
NR
descriptionTRINITY_DN56_c0_g1 5.15×10−6 −17.57 5.30×10−41 1.65×10−36 是 Yes 下调
Down33445.52 0.12 假设蛋白质
Hypothetical proteinTRINITY_DN56_c1_g1 5.39×10−6 −17.50 8.29×10−41 1.65×10−36 是 Yes 下调
Down2710.44 0.01 假设蛋白质
MIMGU_mgv1a026582mg,
hypothetical protein
MIMGU_mgv1a026582mgTRINITY_DN388_c0_g1 1.36×10−5 −16.16 4.43×10−40 5.88×10−36 是 Yes 下调
Down37524.60 0.40 假设蛋白质
Hypothetical proteinTRINITY_DN56_c0_g2 2.62×10−5 −15.22 7.54×10−40 7.52×10−36 是Yes 下调
Down24571.29 0.53 假设蛋白质
Hypothetical proteinTRINITY_DN1368_c0_g1 2.82×10−5 −15.11 8.27×10−39 6.59×10−35 是 Yes 下调
Down16978.15 0.39 未注释 No TRINITY_DN863_c0_g2 2.14×10−5 −15.51 4.02×10−38 2.67×10−34 是 Yes 下调
Down2861.27 0.05 假设蛋白质
MIMGU_mgv1a026582mg hypothetical protein MIMGU_mgv1a026582mgTRINITY_DN1333_c0_g1 2.42×10−5 −15.33 1.41×10−37 8.01×10−34 是 Yes 下调
Down8296.82 0.16 未注释 No TRINITY_DN893_c0_g2 2.93×10−6 −18.38 3.72×10−35 1.86×10−31 是 Yes 下调
Down11335.30 0.00 假设蛋白质
Hypothetical proteinTRINITY_DN4426_c0_g1 8.51×10−5 −13.52 7.01×10−35 3.11×10−31 是 Yes 下调
Down31665.60 4.91 假设蛋白质
Hypothetical proteinTRINITY_DN6139_c0_g1 6.48×10−5 −13.91 4.83×10−34 1.93×10−30 是 Yes 下调
Down17471.00 0.87 外壳蛋白
Coat protein表 5 部分差异表达基因GO富集结果
Table 5. GO enrichment results of some differentially expressed genes
富集到该GO term的基因
转录本数目
Number of genes enriched to the GO termGO Term对应
的编号
ID Corresponding
to Go termGO三大分类
Three categories
of GOGO功能描述
Go function description该GO在目
标基因集中
占有的比例
The proportion of the GO in the target gene set/%该GO在背景基因转录本中占有的比例
The proportion of the GO in the background gene/%未经校正
的P值
P value
uncorrected校正后
的P值
P value
corrected37 GO:0042744 生物过程 BP 过氧化氢分解过程
Hydrogen peroxide catabolic process0.62 0.35 0.00 0.00 37 GO:0042743 生物过程 BP 过氧化氢代谢过程
Hydrogen peroxide metabolic process0.62 0.35 0.00 0.00 65 GO:0072330 生物过程 BP 一元羧酸生物合成工艺
Monocarboxylic acid biosynthetic process1.09 0.75 0.00 0.02 48 GO:0000272 生物过程 BP 多糖分解代谢过程
Polysaccharide catabolic process0.80 0.54 0.00 0.04 54 GO:0030001 生物过程 BP 金属离子输运
Metal ion transport0.91 0.62 0.00 0.04 128 GO:0005576 细胞组分 CC 细胞外区
Extracellular region2.15 1.45 0.00 0.00 73 GO:0005618 细胞组分 CC 细胞壁 Cell wall 1.23 0.82 0.00 0.00 73 GO:0030312 细胞组分 CC 外部封装结构
External encapsulating structure1.23 0.82 0.00 0.00 2136 GO:0031224 细胞组分 CC 膜的固有成分
Intrinsic component of membrane35.88 34.47 0.00 0.15 37 GO:0031226 细胞组分 CC 质膜固有成分
Intrinsic component of plasma membrane0.62 0.43 0.00 0.18 209 GO:0001071 分子功能 MF 核酸结合转录因子活性
Nucleic acid binding transcription factor activity3.51 2.48 0.00 0.00 209 GO:0003700 分子功能 MF 序列特异性DNA结合转录因子活性
Transcription factor activity, sequence-specific DNA binding3.51 0.48 0.00 0.00 118 GO:0004497 分子功能 MF 单加氧酶活性
Monooxygenase activity1.98 1.27 0.00 0.00 137 GO:0005506 分子功能 MF 铁离子结合
Iron ion binding2.30 1.54 0.00 0.00 147 GO:0020037 分子功能 MF 血红素结合
Heme binding2.47 1.69 0.00 0.00 表 6 差异表达基因KEGG富集部分分析
Table 6. Partial results of analysis on KEGG enrichment of differentially expressed genes
基因数目
Gene number通路编号
Pathway ID描述
Description该KEGG在目标基因
集中占有的比例
The proportionof KEGG in
target gene concentration/%该KEGG在背景中
占有的比例
The proportion of the
KEGG in the background/%未经校正的P值
P value uncorrected校正后的P值
P value corrected78 map00940 苯丙酸生物合成
Phenylpropanoid biosynthesis2.71 1.36 0.00 0.00 26 map00941 类黄酮生物合成
Flavonoid biosynthesis0.90 0.38 0.00 0.00 17 map00945 二苯乙烯类、二芳基庚烷类和姜辣素的生物合成
Stilbenoid, diarylheptanoid and gingerol biosynthesis0.59 0.26 0.00 0.00 71 map04016 MAPK信号通路-植物
MAPK signaling pathway-plant2.47 1.67 0.00 0.00 63 map00500 淀粉和蔗糖代谢
Starch and sucrose metabolism2.19 1.52 0.00 0.01 26 map00350 酪氨酸代谢
Tyrosine metabolism0.90 0.52 0.00 0.01 92 map04075 植物激素信号转导
Plant hormone signal transduction3.20 2.42 0.00 0.01 26 map00040 戊糖和葡萄糖醛酸的相互转化
Pentose and glucuronate interconversions0.90 0.53 0.00 0.01 39 map04146 过氧化物酶体
Peroxisome1.36 0.88 0.00 0.01 28 map00592 α-亚麻酸代谢
alpha-Linolenic acid metabolism0.97 0.60 0.00 0.02 20 map00360 苯丙氨酸代谢
Phenylalanine metabolism0.69 0.40 0.00 0.03 24 map00460 氰胺酸代谢
Cyanoamino acid metabolism0.83 0.50 0.00 0.03 16 map00906 类胡萝卜素生物合成
Carotenoid biosynthesis0.56 0.31 0.00 0.05 16 map00950 异喹啉生物碱的生物合成
Isoquinoline alkaloid biosynthesis0.56 0.31 0.00 0.04 23 map00130 泛醌和其他萜类醌的生物合
Ubiquinone and other terpenoid-quinone biosynthesis0.80 0.50 0.00 0.05 32 map00260 甘氨酸、丝氨酸和苏氨酸代谢
Glycine, serine and threonine metabolism1.11 0.78 0.00 0.06 23 map04712 植物昼夜节律
Circadian rhythm-plant0.80 0.52 0.00 0.06 28 map00051 果糖和甘露糖代谢
Fructose and mannose metabolism0.97 0.65 0.00 0.07 22 map00052 半乳糖代谢
Galactose metabolism0.76 0.50 0.01 0.10 50 map00520 氨基糖和核苷酸糖代谢
Amino sugar and nucleotide sugar metabolism1.74 1.34 0.01 0.10 表 7 20个差异表达基因qRT-PCR的RQ值
Table 7. RQ values of qRT-PCR for 20 differentially expressed genes
基因
Gene正常种
RQ值
RQ value in CK平原种植
退化种 RQ值
RQ value in BZ调节
RegulationRNA-seq调节
Regulation in
RNA-seq抗病反应蛋白 Disease resistance response protein 1.00 b 216.68 a 上调 Up 上调 Up 抗病蛋白 RGA2 Disease resistance protein RGA2 1.00 b 6.29 a 上调 Up 上调 Up 乙烯受体2 Ethylene receptor 2 1.00 a 0.48 b 下调 Down 下调 Down 过氧化氢酶1 Catalase 1 1.00 b 8.23 a 上调 Up 上调 Up 蔗糖合成酶 Sucrose synthase 1.00 b 10.50 a 上调 Up 上调 Up 抗病蛋白 RPS5 Disease resistance protein RPS5 1.00 b 10.02 a 上调 Up 上调 Up 抗病蛋白 Disease resistance protein 1.00 b 10.04 a 上调 Up 上调 Up 类烟草病毒增殖蛋白3 Tobamovirus multiplication protein 3-like 1.00 a 0.78 b 下调 Down 下调 Down 植物抗病反应蛋白 Plant disease resistance response protein 1.00 b 10.01 a 上调 Up 上调 Up F-box家族蛋白 F-box family protein 1.00 b 7.815577 a 上调 Up 上调 Up 超氧化物歧化酶 [Cu-Zn] Superoxide dismutase [Cu-Zn] 1.00 b 9.217215 a 上调 Up 上调 Up 逆转录病毒相关 Pol多聚蛋白系-1 Retrovirus-related Pol polyprotein LINE-1 1.00 a 0.13 b 下调 Down 下调 Down 转座子 RE1的逆转录病毒相关 Pol多蛋白 Retrovirus-related Pol polyprotein from transposon RE1 1.00 a 0.01 b 下调 Down 下调 Down 转座子 TNT逆转录病毒相关 Pol多蛋白 Retrovirus-related Pol polyprotein from transposon TNT 1.00 a 0.06 b 下调 Down 下调 Down BZIP转录因子家族蛋白 BZIP transcription factor family protein 1.00 a 0.46 b 下调 Down 下调 Down 过氧化物酶 Peroxidase 1.00 b 16.00 a 上调 Up 上调 Up 脱落酸受体 PYR1 Abscisic acid receptor PYR1 1.00 a 0.25 b 下调 Down 下调 Down 抗病蛋白 RPS2 Disease resistance protein RPS2 1.00 b 5.40 a 上调 Up 上调 Up 生长素反应因子1 Auxin response factor 1 1.00 a 0.50 b 下调 Down 下调 Down 烟草病毒增殖蛋白1 Tobamovirus multiplication protein 1 1.00 a 0.49 b 下调 Down 下调 Down 甘油醛-3-磷酸脱氢酶 GAPDH 1.00 a 1.00 a - - 注:同行数据后不同小写字母表示显著性差异(P<0.05)。
Note: Different lowercase letters in each line indicate significant difference at 0.05 level. -
[1] 余志平, 林海红, 余俊红, 等. 铅山红芽芋产业发展概况 [J]. 长江蔬菜, 2016(20):34−36. doi: 10.3865/j.issn.1001-3547.2016.20.015YU Z P, LIN H H, YU J H, et al. Development of red bud taro industry in Yanshan [J]. Journal of Changjiang Vegetables, 2016(20): 34−36.(in Chinese) doi: 10.3865/j.issn.1001-3547.2016.20.015 [2] 姜绍通, 程元珍, 郑志, 等. 红芽芋营养成分分析及评价 [J]. 食品科学, 2012, 33(11):269−272.JIANG S T, CHENG Y Z, ZHENG Z, et al. Analysis and evaluation of nutritional components of red bud taro (Colocasia esulenla L. Schott) [J]. Food Science, 2012, 33(11): 269−272.(in Chinese) [3] 山娜, 杨文俊, 李斌. 举全县之力, 打造铅山红芽芋支柱产业 [J]. 长江蔬菜, 2016(4):9−10. doi: 10.3865/j.issn.1001-3547.2016.04.005SHAN N, YANG W J, LI B. Take the efforts of the whole county to build the pillar industry of red bud taro [J]. Journal of Changjiang Vegetables, 2016(4): 9−10.(in Chinese) doi: 10.3865/j.issn.1001-3547.2016.04.005 [4] 李火金. 铅山红芽芋茎尖脱毒组培繁育及高产栽培 [J]. 中国蔬菜, 2012(2):45−46.LI H J. Virus free tissue culture and high yield cultivation of the shoot tips of the red bud taro [J]. China Vegetables, 2012(2): 45−46.(in Chinese) [5] 江芹, 廖华俊, 董玲, 等. 红芽芋的丛生芽诱导和再生体系建立 [J]. 分子植物育种, 2015, 13(3):675−679.JIANG Q, LIAO H J, DONG L, et al. Induction of multiple shoot and establishment of regeneration system in Clocasia escalenta schott [J]. Molecular Plant Breeding, 2015, 13(3): 675−679.(in Chinese) [6] 邓接楼, 曹昊玮, 李玲, 等. 脱毒红芽芋试管芋盆栽种植的农艺性状分析 [J]. 分子植物育种, 2019, 17(22):7500−7506.DENG J L, CAO H W, LI L, et al. Analysis on agronomic traits of virus-free test-tube taro planted in pots [J]. Molecular Plant Breeding, 2019, 17(22): 7500−7506.(in Chinese) [7] 胡国君, 董雅凤, 张尊平, 等. 植物类病毒脱除技术进展 [J]. 植物保护学报, 2017, 44(2):177−184.HU G J, DONG Y F, ZHANG Z P, et al. Progress in plant viroid elimination techniques [J]. Acta Phytophylacica Sinica, 2017, 44(2): 177−184.(in Chinese) [8] YANG X X, POPOVA E, SHUKLA M R, et al. Root cryopreservation to biobank medicinal plants: A case study for Hypericum perforatum L [J]. In Vitro Cellular & Developmental Biology-Plant, 2019, 55(4): 392−402. [9] KULUS D, REWERS M, SEROCKA M, et al. Cryopreservation by encapsulation-dehydration affects the vegetative growth of Chrysanthemum but does not disturb its chimeric structure [J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2019, 138(1): 153−166. doi: 10.1007/s11240-019-01614-6 [10] LI W B, HARTUNG J S, LEVY L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with Citrus huanglongbing [J]. Journal of Microbiological Methods, 2006, 66(1): 104−115. doi: 10.1016/j.mimet.2005.10.018 [11] WANG Q C, VALKONEN J P T. Cryotherapy of shoot tips: Novel pathogen eradication method [J]. Trends in Plant Science, 2009, 14(3): 119−122. doi: 10.1016/j.tplants.2008.11.010 [12] HELLIOT B, PANIS B, POUMAY Y, et al. Cryopreservation for the elimination of cucumber mosaic and banana streak viruses from banana (Musa spp.) [J]. Plant Cell Reports, 2002, 20(12): 1117−1122. doi: 10.1007/s00299-002-0458-8 [13] 李涵, 陆琳, 瞿素萍, 等. 杂交兰种苗超低温脱毒技术研究 [J]. 中国农业科技导报, 2018, 20(1):147−153.LI H, LU L, QU S P, et al. Study of hybrid orchid seedlings on virus elimination using cryopreservation technology [J]. Journal of Agricultural Science and Technology, 2018, 20(1): 147−153.(in Chinese) [14] 李艳林, 渠慎春, 栾雨婷, 等. 苹果茎尖超低温脱毒体系的建立 [J]. 分子植物育种, 2019, 17(9):2982−2995.LI Y L, QU S C, LUAN Y T, et al. Establishment of cryopreservation detoxification system of apple shoot-tips [J]. Molecular Plant Breeding, 2019, 17(9): 2982−2995.(in Chinese) [15] 周金鑫, 胡新文, 张海文, 等. ABA在生物胁迫应答中的调控作用 [J]. 农业生物技术学报, 2008, 16(1):169−174. doi: 10.3969/j.issn.1674-7968.2008.01.032ZHOU J X, HU X W, ZHANG H W, et al. Regulatory role of ABA in plant response to biotic stresses [J]. Journal of Agricultural Biotechnology, 2008, 16(1): 169−174.(in Chinese) doi: 10.3969/j.issn.1674-7968.2008.01.032 [16] XANTHOPOULOU A, GANOPOULOS I, PSOMOPOULOS F, et al. De novo comparative transcriptome analysis of genes involved in fruit morphology of pumpkin cultivars with extreme size difference and development of EST-SSR markers [J]. Gene, 2017, 622: 50−66. doi: 10.1016/j.gene.2017.04.035 [17] UNAMBA C I N, NAG A, SHARMA R K. Next generation sequencing technologies: The doorway to the unexplored genomics of non-model plants [J]. Frontiers in Plant Science, 2015, 6: 1074. [18] COSTA V, ANGELINI C, DE FEIS I, et al. Uncovering the complexity of transcriptomes with RNA-seq [J]. Journal of Biomedicine and Biotechnology, 2010, 2010: 1−19. [19] WANG Z, GERSTEIN M, SNYDER M. RNA-Seq: a revolutionary tool for transcriptomics [J]. Nature Reviews Genetics, 2009, 10(1): 57−63. doi: 10.1038/nrg2484 [20] VIEIRA R L, SILVA A L, ZAFFARI G R, et al. Efficient elimination of virus complex from garlic (Allium sativum L.) by cryotherapy of shoot tips [J]. Acta Physiologiae Plantarum, 2014, 37(1): 1−11. [21] WANG B, WANG R R, CUI Z H, et al. Potential applications of cryogenic technologies to plant genetic improvement and pathogen eradication [J]. Biotechnology Advances, 2014, 32(3): 583−595. doi: 10.1016/j.biotechadv.2014.03.003 [22] WANG Q C, PANIS B, ENGELMANN F, et al. Cryotherapy of shoot tips: A technique for pathogen eradication to produce healthy planting materials and prepare healthy plant genetic resources for cryopreservation [J]. Annals of Applied Biology, 2009, 154(3): 351−363. doi: 10.1111/j.1744-7348.2008.00308.x [23] 何广深. 百子莲胚性愈伤组织超低温保存中钙离子的分布变化及逆境应答机制初探 [D]. 上海: 上海交通大学, 2014.HE G S. Distribution change of Ca2+ and stress response mechanism research of Agapanthus praecox embryogenic callus during cryopreservation [D]. Shanghai: Shanghai Jiaotong University, 2014.(in Chinese) [24] 滕进婧, 李梦芸, 郭纯, 等. 冷冻胁迫转金柑MLP2-1基因拟南芥的转录组测序和代谢通路 [J]. 湖南农业大学学报(自然科学版), 2018, 44(4):376−381.TENG J J, LI M Y, GUO C, et al. Transcriptome sequencing and metabolic pathway analysis of transgenic MLP2-1 gene in Arabidopsis under cold stress [J]. Journal of Hunan Agricultural University (Natural Sciences Edition), 2018, 44(4): 376−381.(in Chinese) [25] 苏谦, 安冬, 王库. 植物激素的受体和诱导基因 [J]. 植物生理学通讯, 2008, 44(6):1202−1208.SU Q, AN D, WANG K. Phytohormone receptors and induced genes in plants [J]. Plant Physiology Communications, 2008, 44(6): 1202−1208.(in Chinese) [26] OHRI P, BHARDWAJ R, BALI S G, et al. The common molecular players in plant hormone crosstalk and signaling [J]. Current Protein & Peptide Science, 2015, 16(5): 369−388. [27] 杨家书, 吴畏, 吴友三, 等. 植物苯丙酸类代谢与小麦对白粉病抗性的关系 [J]. 植物病理学报, 1986, 16(3):169−174.YANG J S, WU W, WU Y S, et al. Relation of metabolism of plant phenylalanine and resistance of wheat to powdery mildew [J]. Acta Phytopathologica Sinica, 1986, 16(3): 169−174.(in Chinese) [28] 陈建中, 盛炳成, 刘克均. 苯丙酸类代谢与苹果对轮纹病抗性的关系 [J]. 果树科学, 1986, 16(3):169−174.CHEN J Z, SHENG B C, LIU K J, et al. The relation between metabolism of phenylalanine and resistance to Physalospora piricola Nose in apple trees [J]. Acta Phytopathologica Sinica, 1986, 16(3): 169−174.(in Chinese) [29] DUAN P G, RAO Y C, ZENG D L, et al. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice [J]. The Plant Journal, 2014, 77(4): 547−557. doi: 10.1111/tpj.12405 [30] LIU S Y, HUA L, DONG S J, et al. OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production [J]. The Plant Journal, 2015, 84(4): 672−681. doi: 10.1111/tpj.13025 [31] LI Y B, FAN C C, XING Y Z, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice [J]. Nature Genetics, 2011, 43(12): 1266. doi: 10.1038/ng.977 [32] 苑智华, 何秀丽, 徐哲, 等. 唐菖蒲球茎形成期蔗糖和淀粉代谢及其相关酶活性 [J]. 林业科学, 2008, 44(8):47−51. doi: 10.3321/j.issn:1001-7488.2008.08.008YUAN Z H, HE X L, XU Z, et al. Metabolism and related enzymes activities of sucrose and starch in the stages of bulb formation of Gladiolus hybridus [J]. Scientia Silvae Sinicae, 2008, 44(8): 47−51.(in Chinese) doi: 10.3321/j.issn:1001-7488.2008.08.008 [33] 孙红梅, 何玲, 王微微, 等. IBA与GA3调控百合鳞片扦插繁殖的 “淀粉-蔗糖” 代谢机制 [J]. 中国农业科学, 2011, 44(4):798−806. doi: 10.3864/j.issn.0578-1752.2011.04.018SUN H M, HE L, WANG W W, et al. Mechanism of starch-sucrose metabolism regulated by IBA as well as GA3 during scale cutting propagation in Lilium [J]. Scientia Agricultura Sinica, 2011, 44(4): 798−806.(in Chinese) doi: 10.3864/j.issn.0578-1752.2011.04.018 [34] HU J P, BAKER A, BARTEL B, et al. Plant peroxisomes: Biogenesis and function [J]. The Plant Cell, 2012, 24(6): 2279−2303. doi: 10.1105/tpc.112.096586 [35] SANDALIO L M, RODRÍGUEZ-SERRANO M, ROMERO-PUERTAS M C, et al. Role of peroxisomes as a source of reactive oxygen species (ROS) signaling molecules[C]//Peroxisomes and Their Key Role in Cellular Signaling and Metabolism, 2013: 231-225. [36] MITTLER R, VANDERAUWERA S, SUZUKI N, et al. ROS signaling: The new wave? [J]. Trends in Plant Science, 2011, 16(6): 300−309. doi: 10.1016/j.tplants.2011.03.007 [37] 于妍, 宋万坤, 刘春燕, 等. 植物天冬氨酸代谢途径关键酶基因研究进展 [J]. 生物技术通报, 2008(S1):7−11, 17.YU Y, SONG W K, LIU C Y, et al. Research development of key enzymes gene on aspartic acid metabolic pathway in plants [J]. Biotechnology Bulletin, 2008(S1): 7−11, 17.(in Chinese) [38] 马瑞肖, 张慧杰, 李馨慧, 等. 丝氨酸/甘氨酸代谢在肿瘤中的研究进展 [J]. 现代肿瘤医学, 2020, 28(6):1021−1024. doi: 10.3969/j.issn.1672-4992.2020.06.033MA R X, ZHANG H J, LI X H, et al. The progress of serine/Glycine metabolism in tumor [J]. Journal of Modern Oncology, 2020, 28(6): 1021−1024.(in Chinese) doi: 10.3969/j.issn.1672-4992.2020.06.033 -







 下载:
下载: