Whole-genome Identification and Bioinformatics of PnPAL Family in Black Peppers
-
摘要:
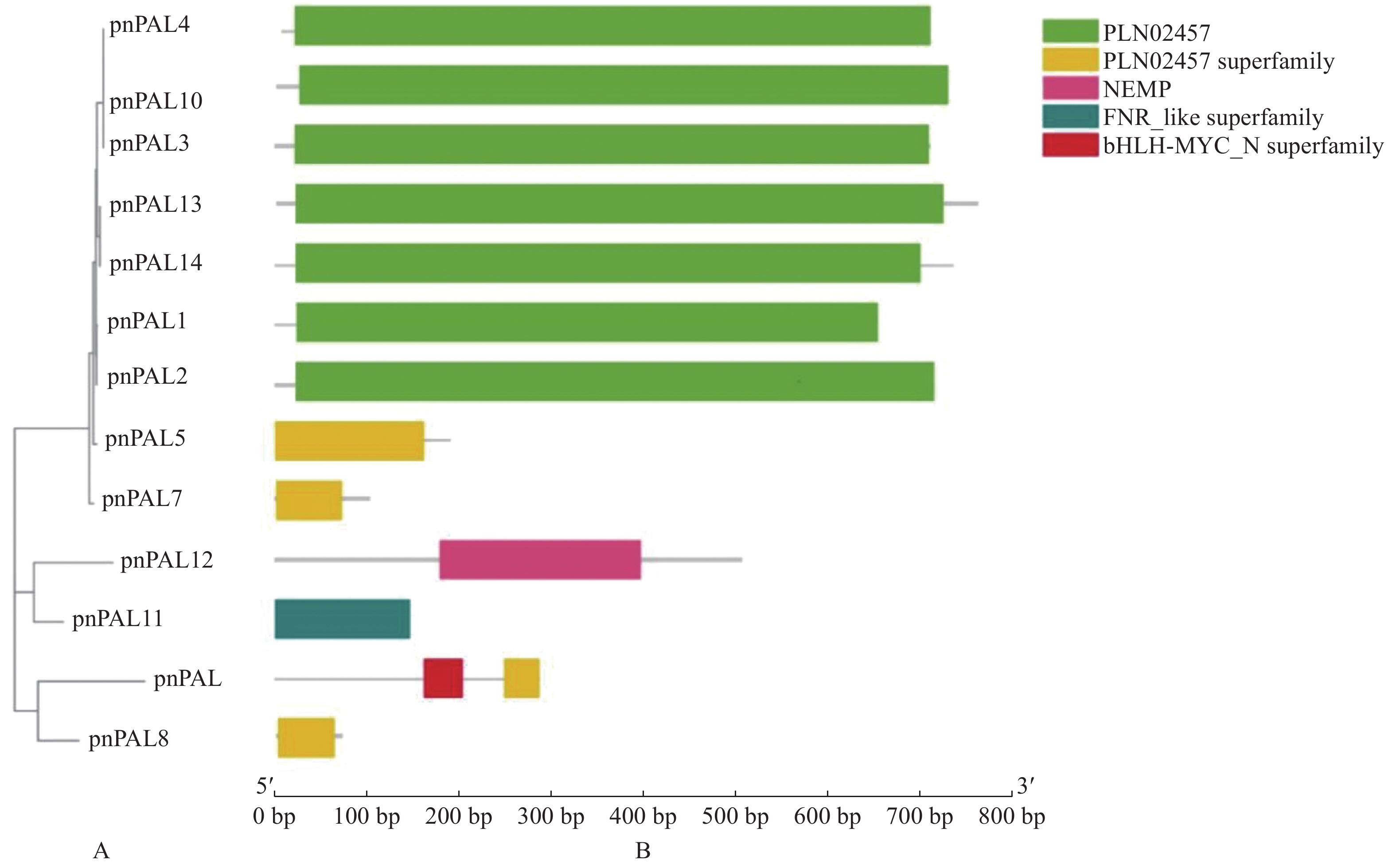
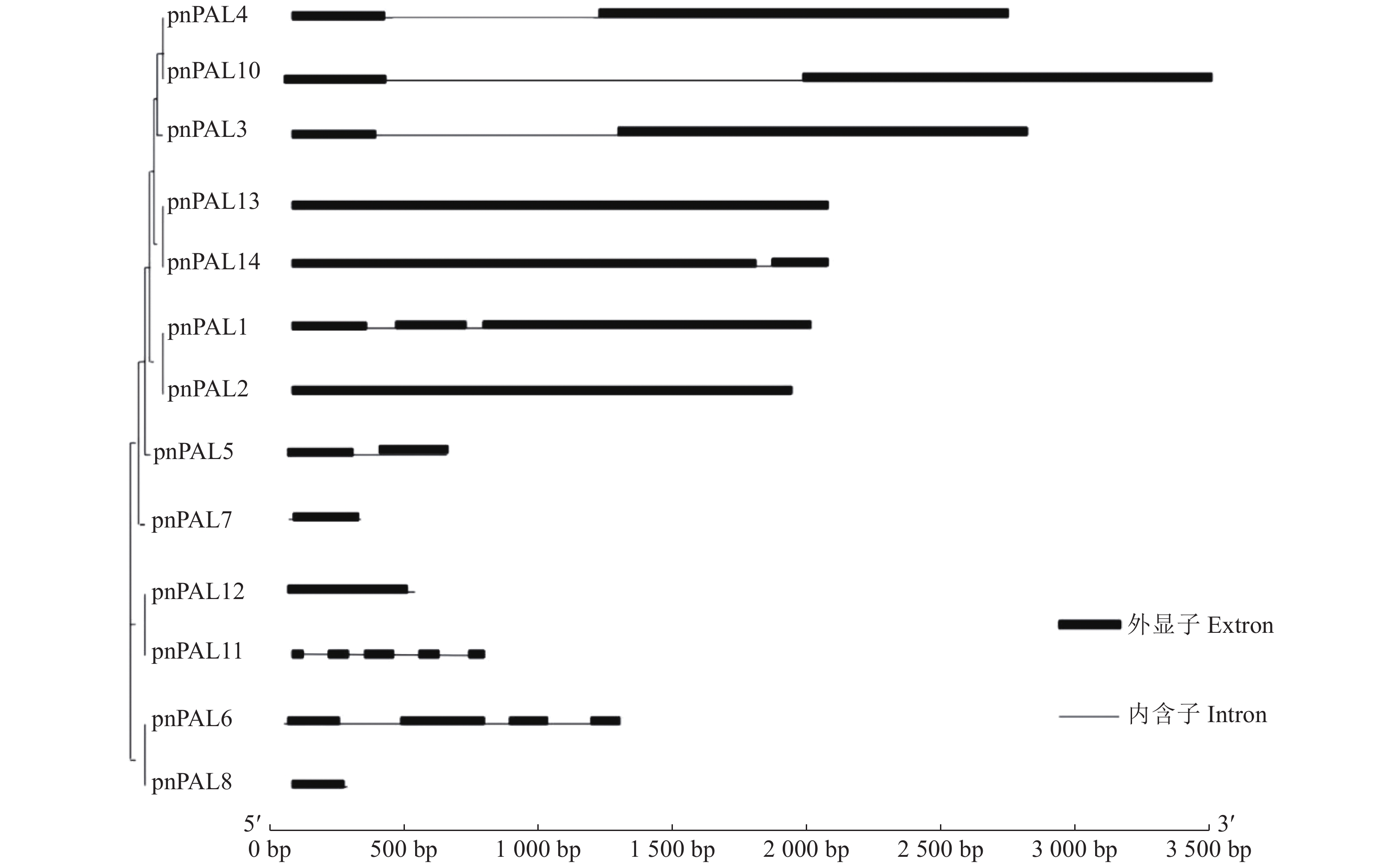
目的 胡椒瘟病是影响胡椒产量的第一大病害,胡椒苯丙氨酸解氨酶(PnPAL)基因是胡椒抗瘟病的关键基因。探明胡椒PnPAL基因家族的基本特征、进化关系及表达模式,以便于其功能研究。 方法 通过全基因组查找、鉴定得到14条胡椒PnPAL家族成员,分别命名为PnPAL1~PnPAL14,并对这14条序列进行生物信息学及表达模式分析。 结果 蛋白质基本信息分析显示,该家族理论等电点在5.76~9.77,分子量在7.374 41~83.431 07 kDa。有7条胡椒的PnPAL基因含有8个motif和PLN02457保守结构域,该7条胡椒PnPAL家族成员被进一步鉴定为胡椒PnPAL家族成员。胡椒PnPAL家族成员含有诸多胁迫响应元件,特别是水杨酸、茉莉酸甲酯等与抵抗病原入侵的顺式作用元件。胡椒的PnPAL基因位于系统发生树基部位置,表明胡椒的PnPAL基因较为古老,与胡椒的系统地位相似。表达模式分析显示,PnPAL10在受病原诱导后基本呈上调趋势,表明PnPAL10可能是胡椒抗瘟病的关键基因。 结论 对胡椒PnPAL基因家族开展了生物信息学及表达模式分析,证明其在抗瘟病中的潜在作用,筛选到关键家族成员PnPAL3。 -
关键词:
- 胡椒 /
- 苯丙氨酸解氨酶(PAL) /
- 生物信息
Abstract:Objective Basic characteristics and evolutional relationship on the phenylpropane metabolic pathway associated with the pepper blast disease that ill-affects the cultivation and production of the globally important spice, black peppers, were studied. Method The phenylpropane metabolic pathway was believed to be key to the blast resistance of black peppers, and phenylalanine ammonialyase (PAL) to be the crucial, rate-limiting enzyme in the pathway. Hence, based on the transcriptomics of a resistant black pepper germplasm the sequences of 14 PALs were identified and named PnPAL1-PnPAL14. Their bioinformatics and expression patterns were analyzed. Result The gene family had a theoretical isoelectric point between pH 5.76 and 9.77 with a molecular weight between 7.37441 kDa and 83.43107 kDa. Seven of the identified PnPALs contained 8 motifs and PLN02457 conserved domains. They belonged to the PnPAL family that consisted of many stress response elements, especially salicylic acid, methyl jasmonate, and other cis-acting elements, which are known to resist the pathogenic invasion. The PnPALs were located at the base of the phylogenetic tree, being relatively old and holding a status like black peppers. PnPAL10 expressed in an up-regulation trend after a pathogenic induction suggesting a close relation of the gene to the blast resistance of the plant. Conclusion A critical role the PnPAL family played in the blast resistance of black peppers was positively identified. -
Key words:
- black pepper /
- phenylalanine ammonialyase /
- biological information
-
图 6 胡椒PnPAL3
基因相对表达分析 注:0 h、8 h、12 h、24 h、48 h表示接种病原菌不同时间点的表达模式;0 h-C、8 h-C、12 h-C、24 h-C、48 h-C 表示接种无菌水不同时间点的表达模式。图中不同小写字母表示接种病原、无菌水基因表达量的显著性水平(P<0.05);图7同此。
Figure 6. Relative expressions of PnPAL3 in black peppers
Note: 0 h, 8 h, 12 h, 24 h, and 48 h are expressions at respective time after pathogenic inoculation; 0 h-C, 8 h-C, 12 h-C, 24 h-C, and 48 h-C, expressions at respective time after sterile water inoculation. Data with different lowercase letters indicate significant difference in expressions between inoculation with pathogens and sterile water (P<0.05). The same as Fig.7.
表 1 实时荧光定量PCR分析所用引物
Table 1. Primers used in RT-PCR analysis
基因名称
Gene name正向引物序列
Forward primer sequence (5′-3′)反向引物序列
Revers primer sequence (5′-3′)PnPAL1 GCAAAGGCCGTGACTTTCG TTCATCACCTCGCAGAAAACG PnPAL2 CGCGGCCAGCTTGAAGAG GCGAGGATGTTAGCGTCGTAGA PnPAL3 CCGACCACTTGACCCATAAGC GGTCCATCTCATGTGTCATTTGC PnPAL4 GCTAGGCCCACTCATAGAAGTGA AGTGTTGTCCATGGACACACCA PnPAL10 CAAAAGCTGAGAAATGTGCTGGT CCGACTTCCCGTTCTCATACTC PnPAL13 ACAACCAGGACGTCAACTCACTG ACGATGTGCTTGACCACCTCTC PnPAL14 GGCACAACTCCAAGGTTACGA GTTGGCGTCGTAGCAAACG 内参序列 GTTGGAGATAGGAGCAAGCAGA TCAAAGACCACCCCATCACCT 表 2 胡椒PnPAL基因家族成员的理化性质
Table 2. Physiochemical properties of PnPAL family in black peppers
名称
Gene name基因编号
Sequence
ID染色体定位
Chromosome
nambur染色体位置
Chromosome
location编码氨基酸个数
AA蛋白质分子量
Mw /kDa等电点
pI亚细胞定位
Subcellular
localizationPnPAL1 Pn2.1119 2 14526770-14528898- 652 70.242 67 5.84 细胞质 Cytoplasm PnPAL2 Pn2.1269 2 15529503-15531650+ 715 76.798 79 5.76 细胞质 Cytoplasm PnPAL3 Pn2.1585 2 18068679-18071787+ 711 77.464 65 5.93 细胞质 Cytoplasm PnPAL4 Pn2.1713 2 19171638-19174695+ 711 77.495 66 5.87 细胞质 Cytoplasm PnPAL5 Pn3.4768 3 37507112-37507766- 191 21.420 58 9.77 细胞核 Cytoplasm PnPAL6 Pn5.1823 5 20214336-20215764- 312 34.470 96 7.75 叶绿体 Chloroplast PnPAL7 Pn6.2505 6 23152346-23152648- 100 10.668 38 8.93 细胞核 Nucleus PnPAL8 Pn6.2507 6 23155823-23156041- 72 7.374 41 8.93 细胞核 Nucleus PnPAL9 Pn6.2506 6 23155093-23155674- 193 20.389 72 7.72 细胞核 Nucleus PnPAL10 Pn8.2617 8 29114372-29118271- 730 79.494 98 6.04 细胞质 Cytoplasm PnPAL11 Pn9.116 9 18656999-18659720- 147 16.374 80 7.77 叶绿体 Chloroplast PnPAL12 Pn18.280 18 240046-240597- 507 57.052 64 8.05 内膜系统 Endometrial system PnPAL13 Pn28.318 28 2239516-2241801- 761 83.431 07 6.08 质膜 Plasma membrane PnPAL14 Pn38.78 38 621136-623422+ 736 80.379 55 6.08 质膜 Plasma membrane -
[1] 刘进平, 郑成木, 郑服丛. 胡椒瘟病与辣椒疫霉 [J]. 热带农业科学, 2001, 21(5):57−61. doi: 10.3969/j.issn.1009-2196.2001.05.014LIU J P, ZHENG C M, ZHENG F C. Black pepper foot rot and Phytophthora capsica [J]. Chinese Journal of Tropical Agriculture, 2001, 21(5): 57−61.(in Chinese) doi: 10.3969/j.issn.1009-2196.2001.05.014 [2] 桑利伟, 刘爱勤, 谭乐和, 等. 海南省胡椒瘟病病原鉴定及发生规律 [J]. 植物保护, 2011, 37(6):168−171. doi: 10.3969/j.issn.0529-1542.2011.06.034SANG L W, LIU A Q, TAN L H, et al. Pathogen identification and occurrence of the pepper Phytophthora foot rot in Hainan Province [J]. Plant Protection, 2011, 37(6): 168−171.(in Chinese) doi: 10.3969/j.issn.0529-1542.2011.06.034 [3] YUAN W, JIANG T, DU K T, et al. Maize phenylalanine ammonia-lyases contribute to resistance to sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation [J]. Molecular Plant Pathology, 2019, 20(10): 1365−1378. doi: 10.1111/mpp.12817 [4] 王敬文, 薛应龙. 植物苯丙氨酸解氨酶的研究——Ⅱ 苯丙氨酸解氨酶在抗马铃薯晚疫病中的作用 [J]. 植物生理学报, 1982(1):35−43.WANG J W, XUE Y L. Study on plant phenylalanine ammonia lyase—Ⅱ the role of phenylalanine ammonia lyase in resistance to potato late blight [J]. Plant Physiology Communications, 1982(1): 35−43.(in Chinese) [5] DIXON R. A Stress-induced phenylpropanoid metabolism [J]. Plant Cell, 1995, 7(7): 1085. doi: 10.2307/3870059 [6] 康晓慧, 雷桅, 张梅. 水稻苯丙氨酸解氨酶的生物信息学分析 [J]. 湖南师范大学自然科学学报, 2010, 33(4):89−94. doi: 10.3969/j.issn.1000-2537.2010.04.018KANG X H, LEI W, ZHANG M. Bioinformatic analysis of phenylalanine ammonia-lyase gene in Oryza sativa [J]. Journal of Natural Science of Hunan Normal University, 2010, 33(4): 89−94.(in Chinese) doi: 10.3969/j.issn.1000-2537.2010.04.018 [7] 杨郁文, 李双, 黄俊宇, 等. 陆地棉苯丙氨酸解氨酶家族基因的鉴定及分析 [J]. 分子植物育种, 2017, 15(4):1184−1191.YANG Y W, LI S, HUANG J Y, et al. Identification and analysis of the gene family of phenylalanine ammonia-lyase in upland cotton [J]. Molecular Plant Breeding, 2017, 15(4): 1184−1191.(in Chinese) [8] 范丽. 桑树木质素合成基因的生物信息和功能分析[D].重庆: 西南大学, 2013.FAN L. Biological information and function analysis of mulberry lignin synthesis gene[D]. Chongqing: Southwest University, 2013. [9] 宋婕. 丹参苯丙氨酸解氨酶基因(SmPAL1)的克隆及其功能初探[D]. 西安: 陕西师范大学, 2007.SONG J. Molecular cloning of a phenylalanine ammonia—lyase gene(SmPALl) from Salvia miltiorrhiza and the primary study on its function[D]. Xi'an: Shaanxi Normal University, 2007.(in chinese) [10] 冯立娟, 尹燕雷, 焦其庆, 等. 石榴PAL基因的克隆与表达分析 [J]. 核农学报, 2018, 32(7):1320−1329. doi: 10.11869/j.issn.100-8551.2018.07.1320FENG L J, YIN Y L, JIAO Q Q, et al. Cloning and expression analysis of PAL gene in pomegranate (Punica granatum L.) [J]. Journal of Nuclear Agricultural Sciences, 2018, 32(7): 1320−1329.(in Chinese) doi: 10.11869/j.issn.100-8551.2018.07.1320 [11] 郭鹏飞, 雷健, 罗佳佳, 等. 柱花草苯丙氨酸解氨酶(SgPALs)对生物胁迫与非生物胁迫的响应 [J]. 热带作物学报, 2019, 40(9):1742−1751. doi: 10.3969/j.issn.1000-2561.2019.09.011GUO P F, LEI J, LUO J J, et al. Response of phenylpropane ammonia-lyase on biotic and abiotic stress in Stylosanthes [J]. Chinese Journal of Tropical Crops, 2019, 40(9): 1742−1751.(in Chinese) doi: 10.3969/j.issn.1000-2561.2019.09.011 [12] 雒军, 王引权, 温随超, 等. 当归苯丙氨酸解氨酶基因片段克隆和组织特异性表达分析 [J]. 草业学报, 2014, 23(4):130−137. doi: 10.11686/cyxb20140416LUO J, WANG Y Q, WEN S C, et al. Cloning and tissue-specific expression analysis of phenylalanine ammonia-lyase gene fragment in Angelica sinensis [J]. Acta Prataculturae Sinica, 2014, 23(4): 130−137.(in Chinese) doi: 10.11686/cyxb20140416 [13] HAO C, XIA Z, FAN R, et al. De novo transcriptome sequencing of black pepper (Piper nigrum L.) and an analysis of genes involved in phenylpropanoid metabolism in response to Phytophthora capsici [J]. BMC Genomics, 2016, 17(1): 822. doi: 10.1186/s12864-016-3155-7 [14] 文印. 基部被子植物水力结构进化及其与光合的关联[D].南宁: 广西大学, 2019.WEN Y. Evolution of hydraulic structure in basal angiosperms and its relation to photosynthesis - case studies[D]. Nanning: Guangxi University, 2019. [15] 杨永, 傅德志, 王祺. 被子植物花的起源:假说和证据 [J]. 西北植物学报, 2004(12):2366−2380. doi: 10.3321/j.issn:1000-4025.2004.12.032YANG Y, FU D Z, WANG Q. Origin of flowers:hypotheses and evidence [J]. Acta Botanica Boreali-occidentalia Sinica, 2004(12): 2366−2380.(in Chinese) doi: 10.3321/j.issn:1000-4025.2004.12.032 [16] 叶小真, 杨婕, 冯丽贞, 等. 桉树PAL基因克隆及焦枯病菌诱导下的表达分析 [J]. 林业科学研究, 2019, 32(6):99−105.YE X Z, YANG J, FENG L, et al. Cloning of PAL Gene from Eucalyptus and Its Expression underCalonectria pseudoreteaudii Stress [J]. Forest Research, 2019, 32(6): 99−105.(in Chinese) [17] 吴远航, 刘秦, 刘攀道, 等. 木薯苯丙氨酸解氨酶基因的克隆及其对低温胁迫的响应 [J]. 热带作物学报, 2019, 40(3):483−489. doi: 10.3969/j.issn.1000-2561.2019.03.010WU Y H, LIU Q, LIU P D, et al. Cloning of Cassava Phenylalanine Ammonia Lyase Genes and Their Responses to Low Temperature Stress [J]. Chinese Journal of Tropical Crops, 2019, 40(3): 483−489.(in Chinese) doi: 10.3969/j.issn.1000-2561.2019.03.010 [18] 高红胜, 张仁英, 许学文, 等. 黄瓜苯丙氨酸解氨酶基因CsPAL的克隆及响应白粉菌侵染的表达分析 [J]. 分子植物育种, 2019, 17(6):1757−1762. doi: 10.13271/j.mpb.017.001757GAO H S, ZHANG R Y, XU X W, et al. Cloning of CsPAL Gene and Its Expression Analysis in response to Powdery Mildew Infection [J]. Molecular Plant Breeding, 2019, 17(6): 1757−1762.(in Chinese) doi: 10.13271/j.mpb.017.001757 [19] 孙海燕, 全雪丽, 付爽, 等. 拟南芥苯丙氨酸解氨酶(PAL)基因的研究进展 [J]. 延边大学农学学报, 2016, 38(1):88−92.SUN H Y, QUAN X L, FU S, et al. Research progress of Phenylalanine ammonia-lyase (PAL) gene in Arabidopsis thaliana [J]. Journal of Agricultural Science Yanbian University, 2016, 38(1): 88−92.(in Chinese) [20] 方长旬, 王清水, 余彦, 等. 不同胁迫条件下化感与非化感水稻PAL多基因家族的差异表达 [J]. 生态学报, 2011, 31(16):4760−4767.FANG C X, WANG Q S, YU Y, et al. Differential expression of PAL multigene family in allelopathic rice and its counterpart exposed to stressful conditions [J]. Acta Ecologica Sinica, 2011, 31(16): 4760−4767.(in Chinese) [21] 王燕, 石强, 薛志东. 蛋白质结构域划分方法及在线服务综述 [J]. 广州大学学报(自然科学版), 2019, 18(1):20−29.WANG Y, SHI Q, XUE Z D. a review of protein domain partitioning methods and online services [J]. Journal of Guangzhou University (Natural Science Edition), 2019, 18(1): 20−29.(in Chinese) [22] DE GRASSI A, LANAVE C, SACCONE C. Genome duplication and gene-family evolution: The case of three OXPHOS gene families [J]. Gene, 2008, 421(1/2): 1−6. [23] WHETTEN R W, SEDEROFF R R. Phenylalanine ammonia-lyase from loblolly pine: Purification of the enzyme and isolation of complementary DNA clones [J]. Plant Physiology, 1992, 98(1): 380−386. doi: 10.1104/pp.98.1.380 [24] YONA A H, MANOR Y S, HERBST R H, et al. Chromosomal duplication is a transient evolutionary solution to stress [J]. PNAS, 2012, 109(51): 21010−21015. doi: 10.1073/pnas.1211150109 [25] 张古文, 刘莉莉, 王显瑞, 等. 谷子HSP70基因家族的全基因组鉴定及生物信息学分析 [J]. 浙江农业学报, 2015, 27(7):1127−1133. doi: 10.3969/j.issn.1004-1524.2015.07.03ZHANG G W, LIU L L, WANG X R, et al. Genome-wide identification and bioinformatics analysis of HSP70 genes in foxtail millet [J]. Acta Agriculturae Zhejiangensis, 2015, 27(7): 1127−1133.(in Chinese) doi: 10.3969/j.issn.1004-1524.2015.07.03 [26] 李文燕. 植物OPR基因家族系统发育分析及水稻OPR家族基因分子生物学功能研究[D]. 广州: 中山大学, 2010.LI W Y. Phylogenetic analysis of OPR gene fam ily in plants and study of molecular biological functions of OPR family genes in rice[J]. Guangzhou: Sun Yat-sen University, 2010. [27] OHTA T. Evolution and Variation of Multigene Families[M]//Evolution and variation of multigene families/. Springer-Verlag, 1980. [28] BAJIC V B, BRENT M R, BROWN R H, et al. Performance assessment of promoter predictions on ENCODE regions in the EGASP experiment [J]. Genome Biology, 2006, 7(S1): S3.1−S313. -







 下载:
下载: