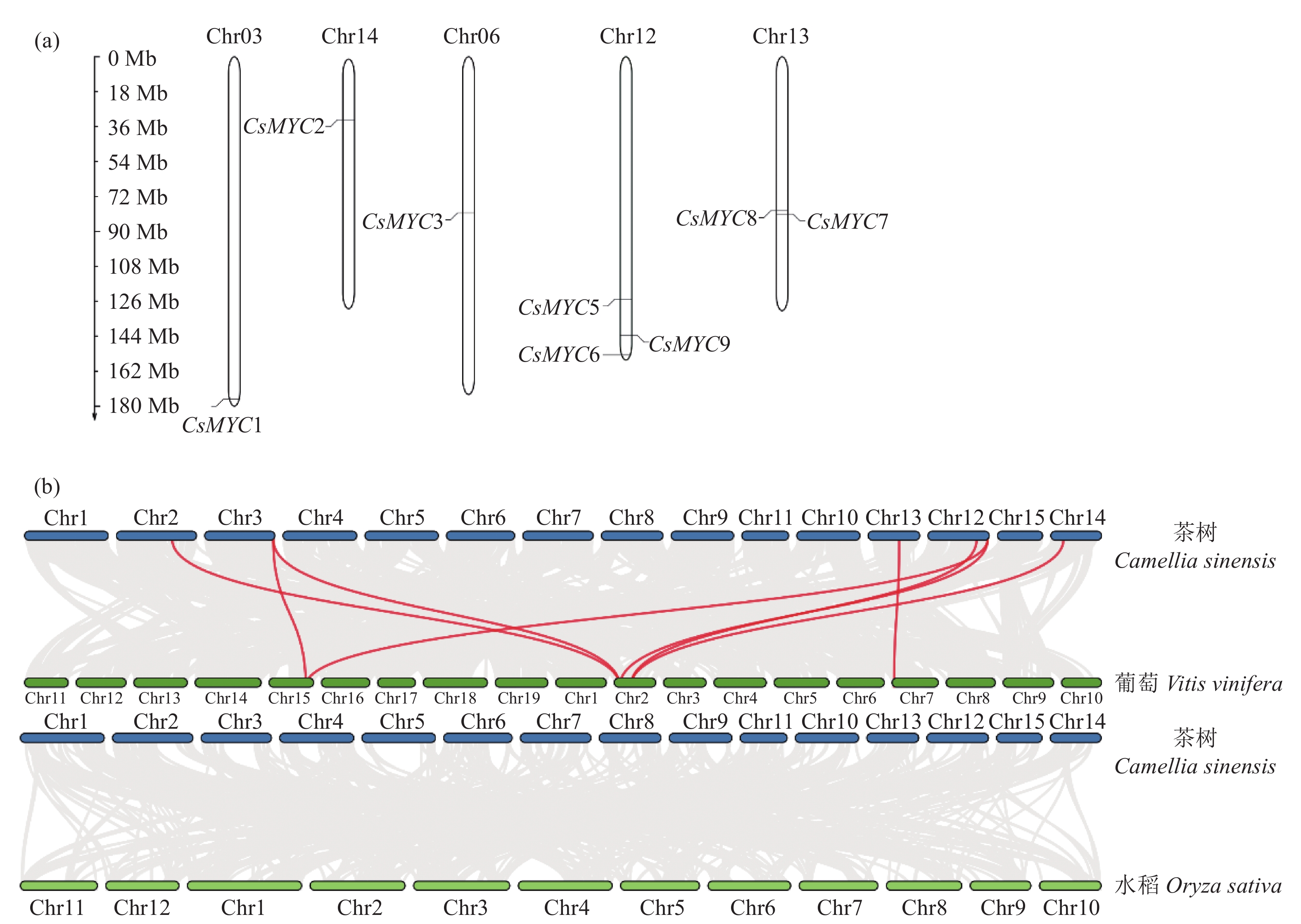
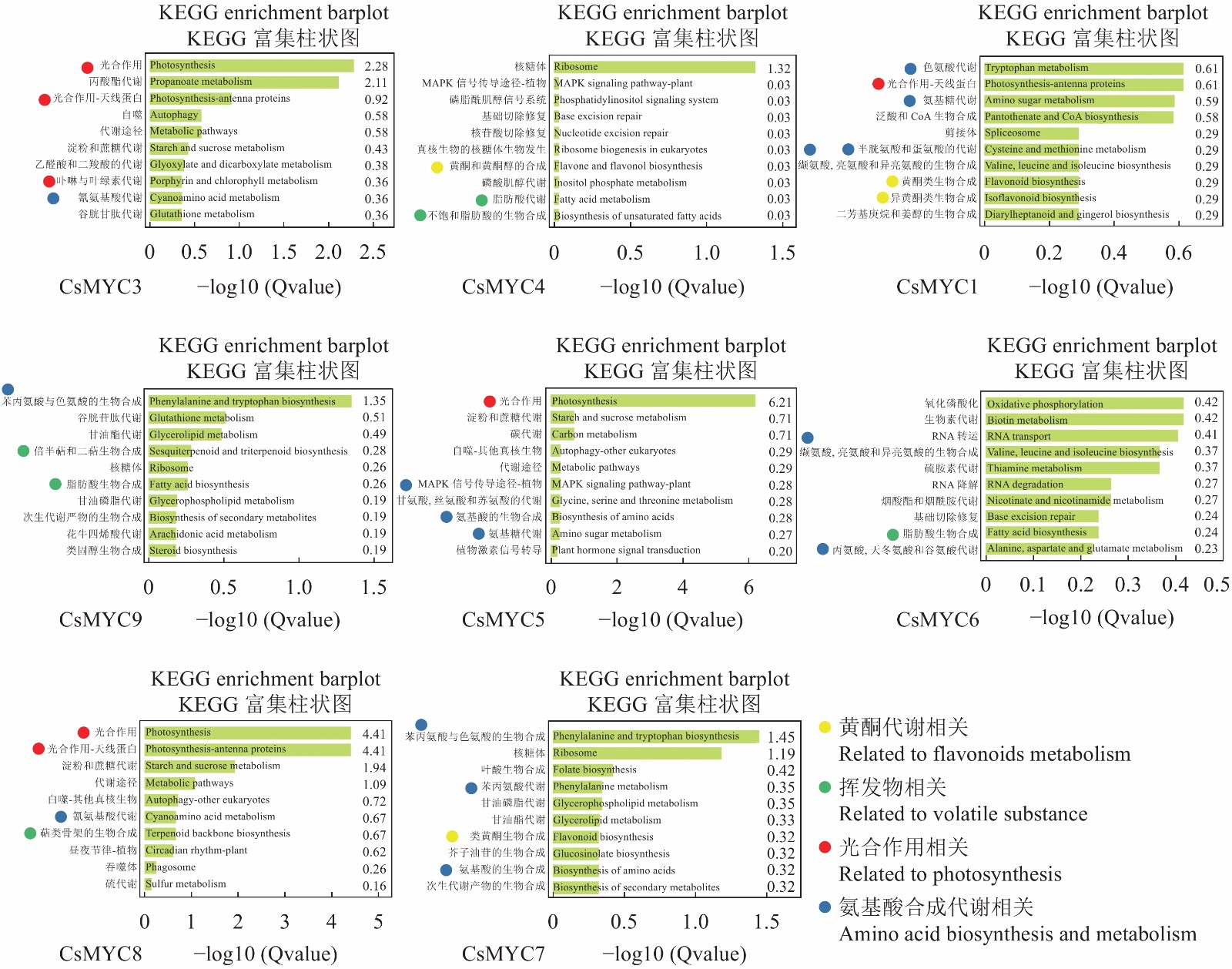
Genome-wide Analysis and Expression Pattern of MYC Family in Camellia sinensis
-
摘要:
目的 髓细胞组织增生蛋白(Myelocytomatosis proteins,MYC)是植物茉莉酸信号转导途径中的重要转录因子。鉴定并分析茶树MYC转录因子有助于了解其潜在的分子机制。 方法 采用生物信息学方法,对茶树CsMYC转录因子进行全基因组范围内的鉴定与分析。 结果 从茶树基因组中总共鉴定出9个CsMYC成员,分布在茶树的5条染色体上。系统进化分析表明植物MYC家族可能起源于陆生植物并发生了谱系特异性分化事件。结构分析表明该家族内含子数目为0~3个,说明该家族在进化过程中发生内含子丢失事件。茶树8个MYC成员能在双子叶植物葡萄中找到同源基因。茶树不同组织转录组分析表明,除了CsMYC2和CsMYC9外,其他成员在芽和叶中表达量较高;荧光定量结果表明,所有成员均响应茉莉酸胁迫处理,同时个别成员也响应干旱、低温和赤霉素处理。使用皮尔森相关系数对与CsMYCs各成员表达量显著相关的基因集进行KEGG功能富集分析表明CsMYCs广泛参与了茶树次级代谢相关途径。 结论 本研究共鉴定出9个CsMYC成员,分析并预测了其结构及潜在分子功能。结合实时荧光定量和功能富集分析表明CsMYC家族在茶树非生物胁迫响应中扮演了重要角色。 Abstract:Objective Myelocytomatosis proteins (MYCs), the key transcription factor in the jasmonic acid signal transduction pathway, in Camellia sinensis were identified and analyzed to help understand the underlying molecular mechanism associated with resistance of tea plants to abiotic stress. Method A genome-wide analysis on the MYC family in tea plant (CsMYC) was conducted using bioinformatics methods. Result Nine CsMYC members were identified in the tea plant genome which distributed unevenly in 5 chromosomes. The phylogenetic analysis suggested that the CsMYCs might originate from terrestrial plants and have undergone lineage specific differentiation. Structurally, the family had only 0 to 3 introns indicating a deletion might have occurred in evolution. Eight of the 9 CsMYCs had homologous genes of dicotyledonous plants like grape, but not of monocotyledonous plants like rice. The transcriptome data showed that, except for CsMYC 2 and CsMYC 9, all members were highly expressed in the buds and leaves. On the fluorescence quantitative tests, all members responded to MeJA stress and some also to drought, low temperature, and/or GA treatments. It indicated a likely close association of tea plants to abiotic stress played by CsMYCs. The KEGG enrichment of the CsMYCs suggested its wide involvement in the secondary metabolism pathways in tea plants. Conclusion Nine CsMYCs in C. sinensis were identified in this study. The structures and potential molecular functions of the members were analyzed and predicted. Combining the results obtained by the real-time fluorescence quantification and functional enrichment analyses, it appeared that the CsMYC family might be closely associated with the response of tea plants to abiotic stress. -
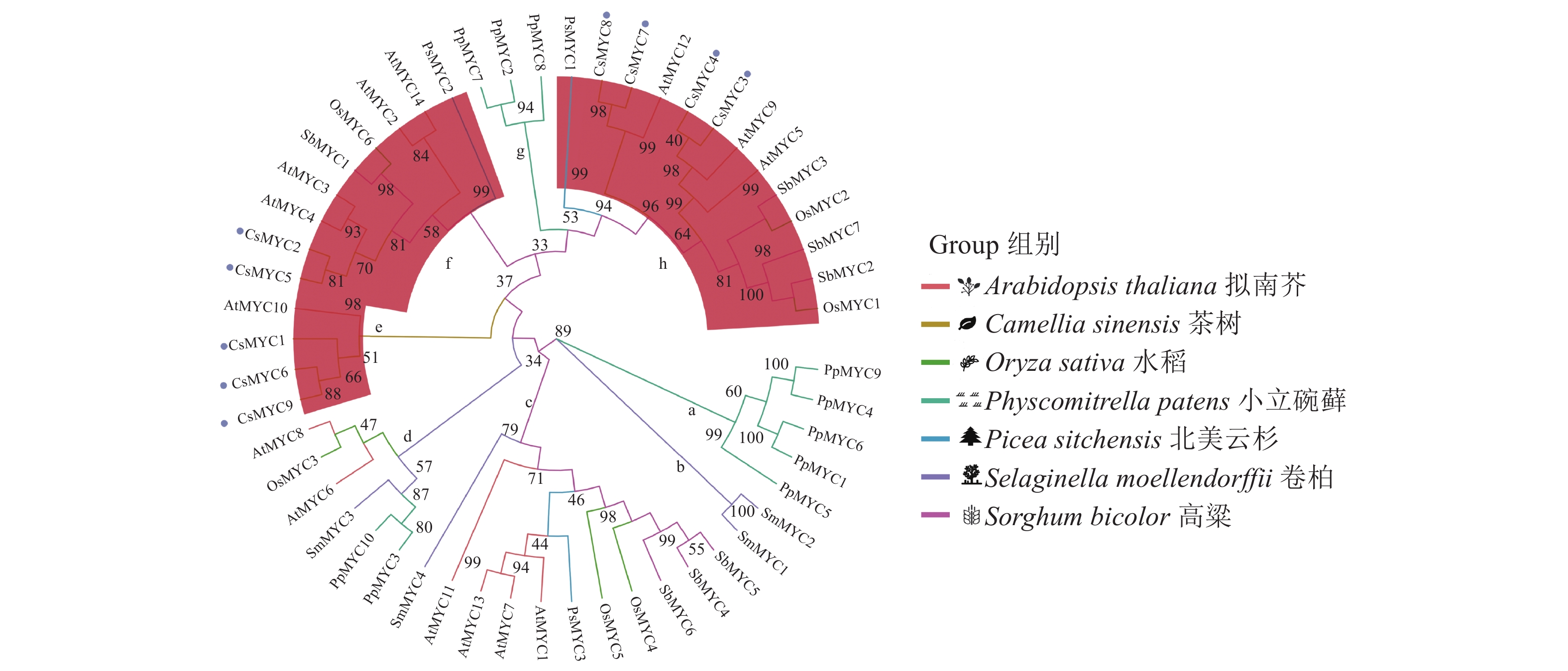
图 2 CsMYC成员的基因结构和保守结构域分析
注:a,CsMYC成员的系统进化树使用邻接法构建;不同颜色方块和数字表示不同模体,其中小号方块(标“*” )表示MEME预测出可能有潜在功能的模体。b,CsMYC成员基因结构图。CDS表示可编码序列,UTR表示非翻译区。c,CsMYC家族成员多序列比对,其中同一列相同颜色方块表示相同氨基酸,左边为bHLH-MYC_N保守结构域,右边为HLH保守结构域。
Figure 2. Structures and conserved domains of CsMYCs
Note: a, phylogenetic tree of CsMYCs calculated using neighbor-joining method; different colored squares and numbers indicate different motifs, and small squares with * symbol represent motifs predicted on MEME website to have potential functions. b, structures of CsMYCs; CDS: codon sequence. UTR: untranslated regions. c, multiple sequence alignment of CsMYCs, where column with same colored squares indicates same amino acid; conserved domain bHLH-MYC N on the left and HLH on the right.
表 1 茶树CsMYC家族成员引物序列
Table 1. Primer sequence of CsMYCs in tea plant
基因名称 Gene names 上游引物 Forward primer 下游引物 Reverse primer CsMYC1 TTGTTGGGTCCGATGCAATG TGGTAAACCGGCAATTCGAG CsMYC2 AACGCCTTCAAGCCCTAATC TTGCGCTTGGCTTTGTCTTC CsMYC3 ATCGGCTTCCATTCCCAAAC ATTGTTTGCCGGCTTCACAC CsMYC4 ATGCATTGCGAGCTGTTGTG TGTGCTCCCCAATTTTTCCC CsMYC5 AAGACAAAGGCAAGCGCAAG AGAACCTTTTTGCGGTGCAG CsMYC6 TTCAAAGCCGAAGCGAATGG TGTTTGGTGCCGCGAAATTG CsMYC7 ATTTGGGGTGATGGGCATTG GCGTGAAGCTTCTGCAAAAC CsMYC8 AGTCGAAATCGGGCAATTCG GCGTGAAGCTTCTGCAAAAC CsMYC9 AGACAGCAATGACCGCTTTC TTGTTGGTGGAAAGCTTGGC CsGAPDH TTGGCATCGTTGAGGGTCT CAGTGGGAACACGGAAAGC 表 2 茶树CsMYC家族成员特征
Table 2. Information on CsMYC in tea plant
基因
Gene基因ID
Gene ID氨基酸数
Amino acid/aa分子量
Molecular weight /kD理论等电点
Academic PI亲水性平均水平
Average hydrophilicity亚细胞定位
Subcellular localizationCsMYC1 TEA022746.1 506 56.437 5.75 −0.596 CN CsMYC2 TEA003964.1 648 71.323 5.34 −0.66 CN CsMYC3 TEA012449.1 595 66.032 6.00 −0.425 CN CsMYC4 TEA019380.1 616 68.151 6.28 −0.459 CN CsMYC5 TEA000833.1 1003 110.270 5.83 −0.331 CN CsMYC6 TEA002159.1 489 54.437 6.07 −0.358 Cyt CsMYC7 TEA013248.1 495 54.856 6.37 −0.476 CN CsMYC8 TEA032433.1 488 53.971 6.35 −0.48 CN CsMYC9 TEA033639.1 442 49.394 6.71 −0.365 Chl 注:CN,细胞核;Chl,叶绿体;Cyt,细胞质。
Note: CN, cell nucleus; Chl, chloroplast; Cyt, cytoplasm. -
[1] ZENG L T, WANG X W, LIAO Y Y, et al. Formation of and changes in phytohormone levels in response to stress during the manufacturing process of oolong tea (Camellia sinensis) [J]. Postharvest Biology and Technology, 2019, 157: 110974. [2] DOMBRECHT B, XUE G P, SPRAGUE S J, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis [J]. The Plant Cell, 2007, 19(7): 2225−2245. doi: 10.1105/tpc.106.048017 [3] WASTERNACK C, HAUSE B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany [J]. Annals of Botany, 2013, 111(6): 1021−1058. doi: 10.1093/aob/mct067 [4] CHINI A, MONTE I, ZAMARREÑO A M, et al. An OPR3-independent pathway uses 4, 5-didehydrojasmonate for jasmonate synthesis [J]. Nature Chemical Biology, 2018, 14(2): 171−178. doi: 10.1038/nchembio.2540 [5] TURNER J G, ELLIS C, DEVOTO A. The jasmonate signal pathway [J]. The Plant Cell, 2002, 14(S1): S153−S164. [6] WOLTERS H, JÜRGENS G. Survival of the flexible: Hormonal growth control and adaptation in plant development [J]. Nature Reviews Genetics, 2009, 10(5): 305−317. [7] LAURIE-BERRY N, JOARDAR V, STREET I H, et al. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae [J]. Molecular Plant Microbe Interactions, 2006, 19(7): 789−800. [8] NIU Y J, FIGUEROA P, BROWSE J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis [J]. Journal of Experimental Botany, 2011, 62(6): 2143−2154. doi: 10.1093/jxb/erq408 [9] ZHANG H T, HEDHILI S, MONTIEL G, et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus [J]. The Plant Journal, 2011, 67(1): 61−71. doi: 10.1111/j.1365-313X.2011.04575.x [10] PATT J M, ROBBINS P S, NIEDZ R, et al. Exogenous application of the plant signalers methyl jasmonate and salicylic acid induces changes in volatile emissions from Citrus foliage and influences the aggregation behavior of Asian Citrus psyllid (Diaphorina citri), vector of Huanglongbing [J]. PLoS One, 2018, 13(3): e0193724. doi: 10.1371/journal.pone.0193724 [11] URAO T, YAMAGUCHI-SHINOZAKI K, MITSUKAWA N, et al. Molecular cloning and characterization of a gene that encodes a MYC-related protein in Arabidopsis [J]. Plant Molecular Biology, 1996, 32(3): 571−576. doi: 10.1007/BF00019112 [12] KAMOLSUKYUNYONG W, SUKHAKET W, RUANJAICHON V, et al. Single-feature polymorphism mapping of isogenic rice lines identifies the influence of terpene synthase on brown planthopper feeding preferences [J]. Rice, 2013, 6(1): 1−9. doi: 10.1186/1939-8433-6-1 [13] SHIMIZU T, LIN F Q, HASEGAWA M, et al. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice [J]. Journal of Biological Chemistry, 2012, 287(23): 19315−19325. [14] 庞茜. 玉米ZmMYC7基因的功能分析[D]. 保定: 河北农业大学, 2019.PANG Q. Functional analysis of ZmMYC7 gene in maize[D]. Baoding: Hebei Agricultural University, 2019. (in Chinese) [15] XIA E H, TONG W, HOU Y, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation [J]. Molecular Plant, 2020, 13(7): 1013−1026. [16] EDDY S R. A probabilistic model of local sequence alignment that simplifies statistical significance estimation [J]. PLoS Computational Biology, 2008, 4(5): e1000069. doi: 10.1371/journal.pcbi.1000069 [17] 王鹏杰, 郑玉成, 林浥, 等. 茶树GRF基因家族的全基因组鉴定及表达分析 [J]. 西北植物学报, 2019, 39(3):413−421.WANG P J, ZHENG Y C, LIN Y, et al. Genome-wide identification and expression analysis of GRF gene family in Camellia sinensis [J]. Acta Botanica Boreali-Occidentalia Sinica, 2019, 39(3): 413−421.(in Chinese) [18] TRAPNELL C, ROBERTS A, GOFF L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks [J]. Nature Protocols, 2012, 7(3): 562−578. doi: 10.1038/nprot.2012.016 [19] ANDERS S, PYL P T, HUBER W. HTSeq—a Python framework to work with high-throughput sequencing data [J]. Bioinformatics, 2015, 31(2): 166−169. doi: 10.1093/bioinformatics/btu638 [20] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method [J]. Methods, 2001, 25(4): 402−408. doi: 10.1006/meth.2001.1262 [21] CANNON S B, MITRA A, BAUMGARTEN A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J]. BMC Plant Biology, 2004, 4: 10. doi: 10.1186/1471-2229-4-10 [22] LYONS R, MANNERS J M, KAZAN K. Jasmonate biosynthesis and signaling in monocots: A comparative overview [J]. Plant Cell Reports, 2013, 32(6): 815−827. doi: 10.1007/s00299-013-1400-y [23] GARRIDO-BIGOTES A, FIGUEROA N E, FIGUEROA P M, et al. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening [J]. PLoS One, 2018, 13(5): e0197118. doi: 10.1371/journal.pone.0197118 [24] CHEN S K, ZHAO H Y, LUO T L, et al. Characteristics and expression pattern of myc genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon [J]. Plants, 2019, 8(8): 274. doi: 10.3390/plants8080274 [25] 郝立冬,郭海滨,李明,等.玉米MYC基因家族的结构特点和表达模式分析[J]. 基因组学与应用生物学. http://kns.cnki.net/kcms/detail/45.1369.Q.20200309.0831.002.html.HAO L D, GUO H B, LI M, et al. Characteristics and expression pattern of maize MYC gene family[J]. Genomics and Applied Biology. http://kns.cnki.net/kcms/detail/45.1369.Q.20200309.0831.002.html. (in Chinese) [26] XU G, GUO C, SHAN H, et al. Divergence of duplicate genes in exon-intron structure [J]. PNAS, 2012, 109(4): 1187−1192. doi: 10.1073/pnas.1109047109 [27] 段龙飞. 茉莉酸信号途径上关键基因家族COI/JAZ/MYC分子进化分析[D]. 杨凌: 西北农林科技大学, 2013.DUAN L F. Molecular evolutionary analysis of the key gene families COI/JAZ/MYC in jasmonic acid signaling pathway[D]. Yangling: Northwest A & F University, 2013. (in Chinese) [28] JEFFARES D C, PENKETT C J, BÄHLER J. Rapidly regulated genes are intron poor [J]. Trends in Genetics, 2008, 24(8): 375−378. doi: 10.1016/j.tig.2008.05.006 [29] SONG S S, HUANG H, WANG J J, et al. MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses [J]. Plant and Cell Physiology, 2017, 58(10): 1752−1763. doi: 10.1093/pcp/pcx112 [30] QI T C, HUANG H, SONG S S, et al. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis [J]. The Plant Cell, 2015, 27(6): 1620−1633. doi: 10.1105/tpc.15.00116 [31] CHEN Q, SUN J Q, ZHAI Q Z, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis [J]. The Plant Cell, 2011, 23(9): 3335−3352. doi: 10.1105/tpc.111.089870 [32] OHTA M, SATO A, RENHU N, et al. MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis [J]. Scientific Reports, 2018, 8(1): 11622. doi: 10.1038/s41598-018-29722-x [33] WEI K F, CHEN H Q. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat [J]. BMC Plant Biology, 2018, 18(1): 309. doi: 10.1186/s12870-018-1529-5 [34] OGAWA S, KAWAHARA-MIKI R, MIYAMOTO K, et al. OsMYC2 mediates numerous defence-related transcriptional changes via jasmonic acid signalling in rice [J]. Biochemical and Biophysical Research Communications, 2017, 486(3): 796−803. doi: 10.1016/j.bbrc.2017.03.125 [35] SZÉKELY G, ABRAHÁM E, CSÉPLO A, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis [J]. The Plant Journal, 2008, 53(1): 11−28. [36] KERKEB L, KRÄMER U. The role of free histidine in xylem loading of nickel in Alyssum lesbiacum and Brassica juncea [J]. Plant Physiology, 2003, 131(2): 716−724. doi: 10.1104/pp102.010686 -







 下载:
下载: