Full-length Transcriptomes of Tea Obtained by Oxford Nanopore Technology
-
摘要:
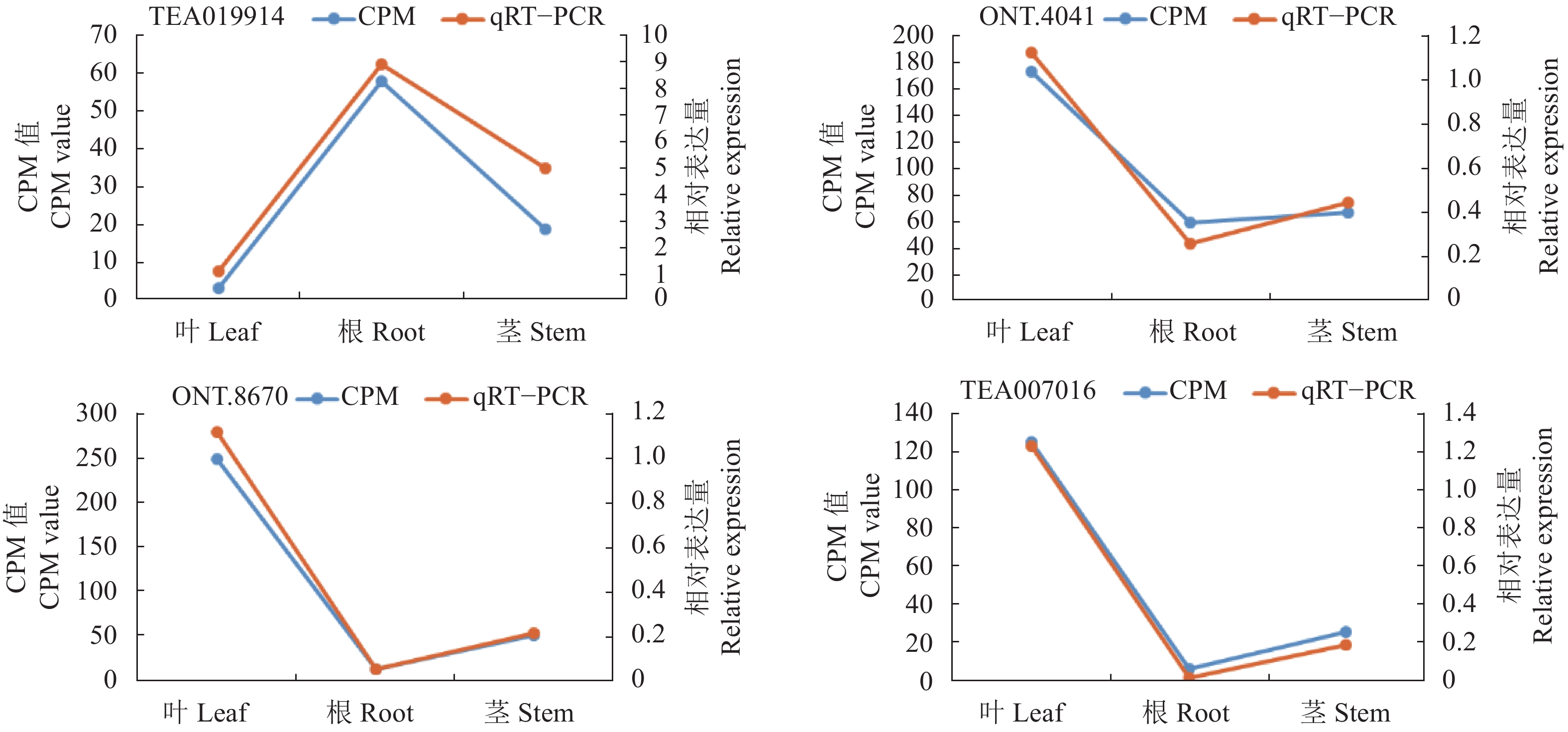
目的 研究茶树的全长转录组测序,以期为解析控制茶叶质量形成的分子机制提供理论依据。 方法 以贵州都匀地区福鼎大白茶叶、根和茎为材料,采用牛津大学的纳米孔技术(Oxford Nanopore Technology, ONT)进行全长转录组测序;将茶树叶、根和茎进行基因表达量分析;为了验证转录组测序结果的可靠性,随机抽取4个基因进行实时荧光定量PCR(qRT-PCR)。 结果 (1)获得69 379条全长转录本、93 102个SSR、45 852个ORF、6 335个转录因子和2 229个lncRNA;(2)获得叶与根、叶与茎以及茎与根的差异转录本,叶与根的差异转录本注释到GO、KEGG和KOG数据库分别有9 646、2 825和7 349个,其中参与茶叶品质形成中茶叶滋味相关代谢途径的有各种氨基酸代谢、类黄酮生物合成和苯丙素生物合成等,与香气相关的代谢途径有泛醌和其他萜烯醌生物合成等;qRT-PCR结果显示其表达量与转录组相一致。 结论 筛选到2个差异基因(ONT.24127.2和TEA003892.1)与谷氨酸合成酶有关,在根部合成茶氨酸后再运输到叶部,进而影响茶叶的品质。研究结果为保障茶叶产量和质量打下良好基础,为后续良种选育和引种及开展后续分子生物学研究提供重要的数据支持。 Abstract:Objective The full-length transcriptomes of tissues of the famed Camellia sinensis (L.) O. Kuntze cv. Fuding Dabaicha were obtained to facilitate cultivar selection in upgrading the Duyun Maojian tea industry and increasing farmers’ income in Guizhou province. Method The Oxford nanopore technology (ONT) was applied to secure the full-length transcriptomes in the leaves, roots, and stems of the tea plant. Gene expressions in the tissues were analyzed. For verification of the ONT results, expressions of 4 randomly selected genes were tested by qRT-PCR. Result The ONT sequencing rendered 69,379 full-length transcripts with approximately 93,102 SSRs, 45,852 ORFs, 6,335 transcription factors, and 2,229 lncRNAs. The differential expression transcripts (DETs) among the leaves, roots, and stems were identified. Between the leaves and roots, there were 9,646 DETs annotated to GO database, 2,825 to KEGG, and 7,348 to KOG. The metabolisms of various amino acids, flavonoid biosynthesis, and phenylpropanol biosynthesis were related to the taste that affects the quality of a tea. Whereas the metabolic pathways involving aroma formation might include biosynthesis of ubiquinone and other terpene quinone. The ONT transcriptomes agreed with the qRT-PCR test results. Conclusion Two differentially expressed genes, ONT.24127.2 and TEA003892.1, were identified to be related to glutamate synthase. Theanine, which is closely associated with tea quality, was synthesized in the roots, then transported to the leaves. The full-length transcriptomes on Fuding Dabaicha demonstrated the practical application of the information for tea cultivar selection. -
Key words:
- Tea plant /
- Fuding Dabaicha /
- Oxford nanopore technology /
- full-length transcriptome
-
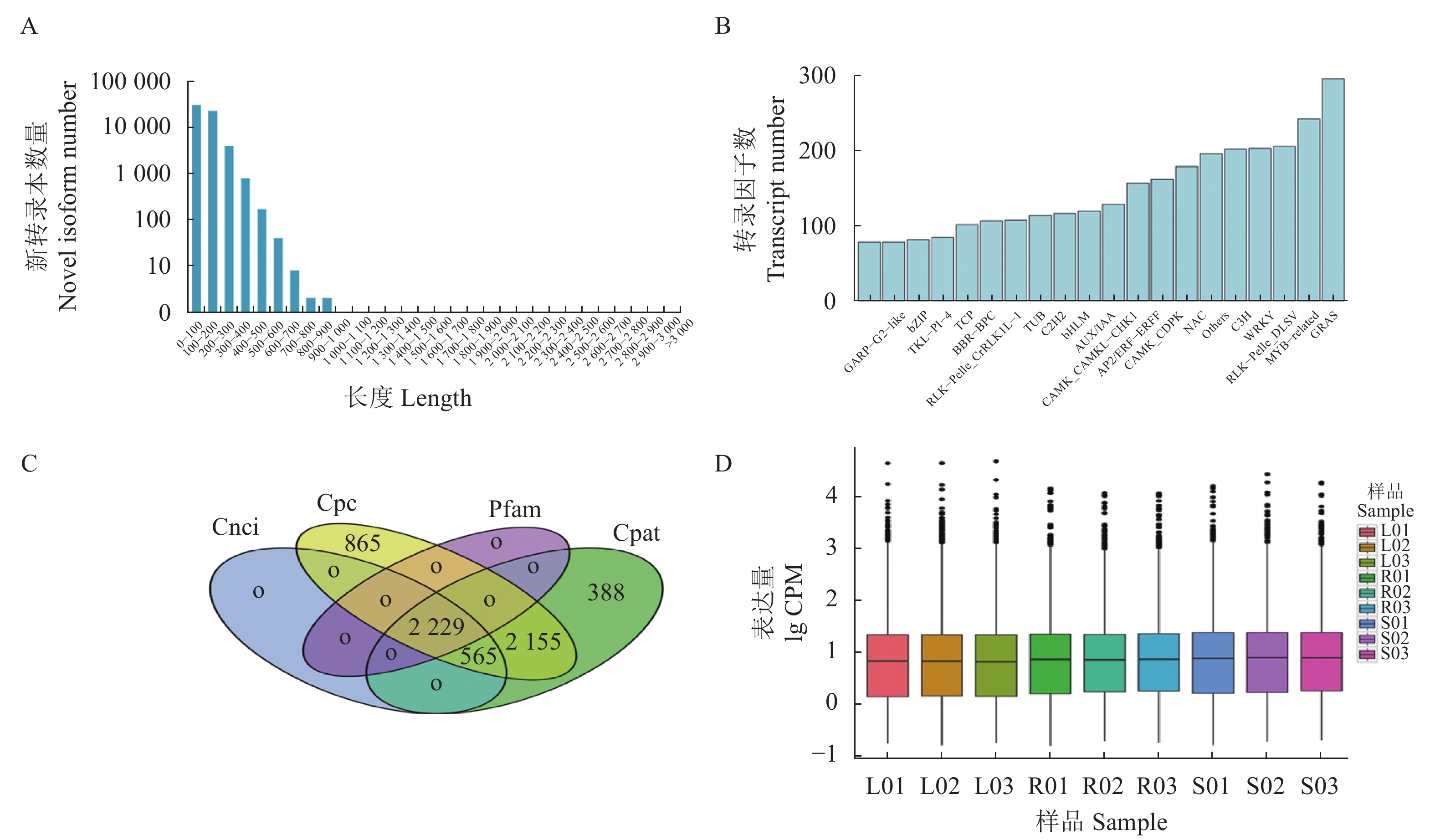
图 1 开放阅读框、转录因子、长链非编码RNA和转录本表达量分析
注:A,ORF编码区氨基酸序列长度分布;B,转录因子种类分布;C,Cnci、Cpc、Pfam、和Cpat四种方法预测的lncRNA韦恩图;D,叶根和茎转录本表达量箱线图。
Figure 1. Analyses on ORF, transcription factor, lncRNA, and transcript expression
Note: A: Distribution of lengths of amino acid sequence in ORF coding region; B: Distribution of transcription factor types; C: Venn graph of lncRNA predicted by 4 methods; D: CPM boxplot of transcripts of leaf, root, and stem.
图 2 叶和根的差异表达转录本GO注释
注:1:代谢过程;2:细胞过程;3:单组织过程;4:生物调节;5:应激反应;6:定位;7:细胞成分组织或生物合成;8:发育过程;9:多细胞生物过程;10:信号;11:生殖;12:生殖过程;13:多组织过程;14:解毒作用;15:生长;16:免疫系统过程;17:节律性过程;18:运动;19:细胞杀伤性;20:生物附着;21:行为;22:细胞;23:细胞组件;24:细胞膜;25:细胞器;26:细胞膜组件;27:细胞器组件;28:大分子复合物;29:细胞膜内腔;30:胞外区;31:细胞连接;32:共质体;33:超分子复合物;34:病毒粒子;35:病毒粒子组件;36:胞外区组件;37:拟核;38:催化活性;39:结合;40:转运活性;41:结构分子活性;42:核酸结合转录因子活性;43:分子功能调节因子;44:信号转导活性;45:抗氧化活性;46:电子载体活性;47:转录因子活性,蛋白结合;48:分子转导活性;49:营养库活性;50:蛋白标签;51:金属伴侣活性;52:翻译调控活性。
Figure 2. GO annotation of DETs between leaf and root
Note: 1: Metabolic process; 2: Cellular process; 3: Single-organism process; 4: Biological regulation; 5: Response to stimulus; 6: Localization; 7: Cellular component organization or biogenesis; 8: Developmental process; 9: Multicellular organismal process; 10: Signaling; 11: Reproduction; 12: Reproductive process; 13: Multi-organism process; 14: Detoxification; 15: Growth; 16: Immune system process; 17: Rhythmic process; 18: Locomotion; 19: Cell-killing; 20: Biological adhesion; 21: Behavior; 22: Cell; 23: Cell part; 24: Membrane; 25: Organelle; 26: Membrane part; 27: Organelle part; 28: Macromolecular complex; 29: Membrane-enclosed lumen; 30: Extracellular region; 31: Cell junction; 32: Symplast; 33: Supramolecular complex; 34: Virion; 35: Virion part; 36: Extracellular region part; 37: Nucleoid; 38: Catalytic activity; 39: Binding; 40: Transporter activity; 41: Structural molecule activity; 42: Nucleic acid binding transcription factor activity; 43: Molecular function regulator; 44: Signal transducer activity; 45: Antioxidant activity; 46: Electron carrier activity; 47: Transcription factor activity, protein-binding; 48: Molecular transducer activity; 49: Nutrient reservoir activity; 50: Protein tag; 51: Metallochaperone activity; 52: Translation regulator activity.
图 3 叶与根差异表达转录本KEGG功能分类
注:1:吞噬体;2:内吞作用;3:过氧化物酶体;4:植物激素信号转导;5:核糖体;6:RNA降解;7:内质网蛋白加工;8:氨酰tRNA生物合成;9:谷胱甘肽代谢;10:光合作用;11:光合生物的固碳作用;12:乙醛酸和二羧酸代谢;13:淀粉和蔗糖代谢;14:苯丙素生物合成;15:氨基酸生物合成;16:碳代谢;17:色氨酸代谢;18:苯丙氨酸、络氨酸和色氨酸生物合成;19:络氨酸代谢;20:缬氨酸、亮氨酸和异亮氨酸降解;21:磷酸肌醇代谢;22:莨菪碱、哌啶和吡啶生物碱的生物合成;23:苯丙氨酸代谢;24:硫代谢;25:氧化磷酸化;26:泛醌和其他萜类醌生物合成;27:类胡萝卜素生物合成;28:抗坏血酸和醛酸代谢;29:戊糖和葡萄糖醛酸转换;30:氮代谢;31:半乳糖代谢;32:果糖和甘露糖代谢;33:氰基氨基酸代谢;34:甘油磷脂代谢;35:α亚麻酸代谢;36:嘧啶代谢;37:脂肪酸降解;38:磷酸戊糖途径;39:类黄酮生物合成;40:萜类化合物生物合成;41:脂肪酸代谢;42:嘌呤代谢;43:卟啉和叶绿素代谢;44:丙酮酸代谢;45:甘氨酸、丝氨酸和苏氨酸代谢;46:光合作用天线蛋白;47:氨基糖和核苷酸糖代谢;48:糖酵解/葡萄糖异生作用;49:半胱氨酸和蛋氨酸代谢;50:植物病原菌互作。
Figure 3. KEGG function annotation of DETs between leaf and root
Note: 1: Phagosome; 2: Endocytosis; 3: Peroxisome; 4: Plant hormone signal transduction; 5: Ribosome; 6: RNA degradation; 7: Protein processing in endoplasmic reticulum; 8: Aminoacyl-tRNA biosynthesis; 9: Glutathione metabolism; 10: Photosynthesis; 11: Carbon fixation in photosynthetic organisms; 12: Glyoxylate and dicarboxylate metabolism; 13: Starch and sucrose metabolism; 14: Phenylpropanoid biosynthesis; 15: Biosynthesis of amino acids; 16: Carbon metabolism; 17: Tryptophan metabolism; 18: Phenylalanine, tyrosine and tryptophan biosysnthesis; 19: Tyrosine metabolism; 20: Valine, leucine and isoleucine degradation; 21: Inositol phosphate metabolism; 22: Tropane, piperidine and pyridine alkaloid biosynthesis; 23: Phenylalanine metabolism; 24: Sulfur metabolism; 25: Oxidative phosphorylation; 26: Ubiquinone and other terpenoid-quinone biosynthesis; 27: Carotenoid biosynthesis; 28: Ascorbate and aldarate metabolism; 29: Pentose and glucuronate interconversions; 30: Nitrogen metabolism; 31: Galactose metabolism; 32: Fructose and mannose metabolism; 33: Cyanoamino acid metabolism; 34: Glycerophospholipid metabolism; 35: Alpha-Linolenic acid metabolism; 36: Pyrimidine metabolism; 37: Fatty acid degradation; 38: Pentose phosphate pathway; 39: Flavonoid biosynthesis; 40: Terpenoid backbone biosynthesis; 41: Fatty acid metabolism; 42: Purine metabolism; 43: Porphyrin and chlorophyll metabolism; 44: Pyruvate metabolism; 45: Glycine, serine and threonine metabolism; 46: Photosynthesis-antenna proteins; 47: Amino sugar and nucleotide sugar metabolism; 48: Glycolysis/gluconeogenesis; 49: Cysteine and methionine metabolism; 50: Plant-pathogen interaction.
表 1 qRT-PCR引物
Table 1. qRT-PCR primer
引物名称
The name of primer引物序列(5′-3′)
The sequence of primer(5′-3′)ONT.4041-1-F TTTGTGGAATGCCCTTTA ONT.4041-1-R GGAAGAATCAATACTGTCAAG ONT.8670-1-F AGGATTGGATGGCGTGTG ONT.8670-1-R GACTCTGCTTGGCTTCTTCA TEA019914.1_gene-1-F AAGTTGTTGCTGATTGATG TEA019914.1_gene-1-R GGTAATGAAGATGGAGTGT TEA007016.1_gene-1-F GCTCATCAGTCAGGTTAT TEA007016.1_gene-1-R GAGGCTGTCTTGTAGTAG GE651107.1 EST1434-1-F(内参基因) CCCTCTCTCCCTTTATCACTCA GE651107.1 EST1434-1-R(内参基因) CCACCAACCGACCAATCC 表 2 Clean data数据
Table 2. Clean data
样品名
Sample IDReads数
The number of
total ReadsClean data平均长度
Mean length of
clean dataClean data N50值
N50 of
clean data全长序列数
Full length
sequence
number全长序列占比
Full length
sequence
proportion比对到参考
转录组Reads个数
The number of
mapped reads比对到参考转录组Reads数目
在全长序列中占的百分比
Mapped ratesL01 6 576 976 1 364 1 503 4 215 412 64.17% 5 609 570 85.29% L02 6 741 658 1 380 1 526 4 819 053 71.55% 6 072 332 90.07% L03 6 273 400 1 393 1 553 4 209 568 67.16% 5 496 900 87.62% R01 7 421 517 1 400 1 605 5 078 770 68.59% 6 132 473 82.63% R02 5 908 300 1 345 1 514 3 879 967 65.74% 5 049 284 85.46% R03 6 290 898 1 452 1 669 4 303 999 68.54% 5 430 183 86.32% S01 7 184 552 1 444 1 667 4 665 036 65.13% 6 003 589 83.56% S02 5 715 955 1 403 1 612 4 045 698 70.84% 5 178 238 90.59% S03 5 616 854 1 412 1 613 3 768 495 67.17% 4 855 092 86.44% 表 3 新转录本注释
Table 3. New isoform annotation
注释数据库
Databases被注释的新转录本数
Annotated new isoform numberCOG 16 999 GO 36 663 KEGG 24 246 KOG 35 099 Pfam 31 029 Swiss-Prot 39 795 eggNOG 56 578 NR 58 139 All 58 398 表 4 差异表达转录本注释
Table 4. Annotation of DETs
差异表达
转录本
DET差异表达
转录本
数目
DET
Number上调
转录本
数目
Up-
regulated下调
转录本
数目
Down-
regulated注释到的
差异表达
转录本
数目
Annotated
DET注释到
COG的
数目
COG注释到
GO的
数目
GO注释到
KEGG的
数目
KEGG注释到
KOG的
数目
KOG注释到
NR的
数目
NR注释到
Pfam的
数目
Pfam注释到
Swiss-Prot的
数目
Swiss-Prot注释到
eggNOG的
数目
eggNOGL01_L02_L03_vs_R01_R02_R03 15 675 7 415 8 260 14 306 5 347 9 649 2 825 7 349 14 166 8 817 10 475 13 859 L01_L02_L03_vs_S01_S02_S03 9 228 4 224 5 004 8 497 3 120 5 865 2 911 4 075 8 423 5 277 6 238 8 243 S01_S02_S03_vs_R01_R02_R03 7 633 4 093 3 540 6 942 2 533 4 739 2 279 3 468 6 876 4 529 5 326 6 688 表 5 部分叶与根差异转录本的KOG分类
Table 5. DETs KOG classification between leaf and root
KOG分类
KOG classificationKOG注释的转录本数
Number of annotated DETs信号转导机制
Signal transduction mechanisms650 能源生产与转换
Energy production and conversion534 无机离子的转运和代谢
Inorganic ion transport and metabolism398 防御机制
Defense mechanisms168 细胞骨架
Cytoskeleton136 辅酶的运输和代谢
Coenzyme transport and metabolism108 RNA加工与修饰
RNA processing and modification91 核苷酸运输与代谢
Nucleotide transport and metabolism80 染色质结构和动力学
Chromatin structure and dynamics30 真核细胞的细胞外结构
Extracellular structures20 核结构 Nuclear structure 6 细胞运动 Cell motility 0 -
[1] 夏丽飞, 孙云南, 宋维希, 等. 基于PacBio平台的紫娟茶树全长转录组分析 [J]. 基因组学与应用生物学, 2020, 39(6):2646−2658.XIA L F, SUN Y N, SONG W X, et al. Full-length transcriptome analysis of Zijuan Tea(Camellia sinensis var. asssamica(Masters) kitamura) base on PacBio platform [J]. Genomics and Applied Biology, 2020, 39(6): 2646−2658.(in Chinese) [2] 谭晓琴, 李伟, 王聪明, 等. ‘紫嫣’和‘紫娟’茶树花青素及主要生化成分季节性的变化 [J]. 热带作物学报, 2021, 42(1):168−174.TAN X Q, LI W, WANG C M, et al. Seasonal changes of anthocyanins and main biochemical components in ‘ziyan’ and ‘zijuan’ tea plants [J]. Chinese Journal of Tropical Crops, 2021, 42(1): 168−174.(in Chinese) [3] 孙云南, 许燕, 冉隆珣, 等. 茶树叶片响应茶饼病侵染的转录组分析 [J]. 茶叶科学, 2020, 40(1):113−124. doi: 10.3969/j.issn.1000-369X.2020.01.013SUN Y N, XU Y, RAN L X, et al. Transcriptome analysis of the tea leaves (Camellia sinensis var. assamica) infected by tea blister blight [J]. Journal of Tea Science, 2020, 40(1): 113−124.(in Chinese) doi: 10.3969/j.issn.1000-369X.2020.01.013 [4] 夏庭君, 吴强盛, 邵雅东, 等. 丛枝菌根真菌对福鼎大白茶生长、侧根数和根系内源激素的影响 [J]. 广西植物, 2018, 38(12):1635−1640. doi: 10.11931/guihaia.gxzw201804030XIA T J, WU Q S, SHAO Y D, et al. Effects of arbuscular mycorrhizal fungi on growth, lateral root number and root endogenous hormones of Camellia sinensis ‘Fuding Dabaicha' [J]. Guihaia, 2018, 38(12): 1635−1640.(in Chinese) doi: 10.11931/guihaia.gxzw201804030 [5] MOLAN A L, DE S, MEAGHER L. Antioxidant activity and polyphenol content of green tea flavan-3-ols and oligomeric proanthocyanidins [J]. International Journal of Food Sciences and Nutrition, 2009, 60(6): 497−506. doi: 10.1080/09637480701781490 [6] ROSS J A, KASUM C M. Dietary flavonoids: Bioavailability, metabolic effects, and safety [J]. Annual Review of Nutrition, 2002, 22: 19−34. doi: 10.1146/annurev.nutr.22.111401.144957 [7] KHAN N, ADHAMI V M, MUKHTAR H. Apoptosis by dietary agents for prevention and treatment of cancer [J]. Biochemical Pharmacology, 2008, 76(11): 1333−1339. doi: 10.1016/j.bcp.2008.07.015 [8] 翟秀明, 唐敏, 李解, 等. 基于RNA-Seq技术的茶树响应高温胁迫转录组差异性分析 [J]. 分子植物育种, 2020, 18(17):5629−5637.ZHAI X M, TANG M, LI J, et al. Difference analysis of heat stress-responsive transcriptome of Camellia sinensis based on RNA-seq technology [J]. Molecular Plant Breeding, 2020, 18(17): 5629−5637.(in Chinese) [9] 廖天悦, 申铁. 基于转录组的福鼎大白茶叶片两种发育阶段的苯丙烷代谢合成途径分析比较 [J]. 贵州师范大学学报(自然科学版), 2021, 39(3):15−22.LIAO T Y, SHEN T. Analysis and comparison of phenylpropane metabolic pathways in two developmental stages of Fuding Dabai tea leaves based on transcriptome [J]. Journal of Guizhou Normal University (Natural Sciences), 2021, 39(3): 15−22.(in Chinese) [10] 高香凤, 王让剑, 刘丰静, 等. 水培枝条取样法在茶树芽叶转录组测序中的可行性鉴定 [J]. 福建农业学报, 2017, 32(2):155−160.GAO X F, WANG R J, LIU F J, et al. Hydroponic culturein sample preparation of tea shoots for RNA sequencing [J]. Fujian Journal of Agricultural Sciences, 2017, 32(2): 155−160.(in Chinese) [11] JANSEN H J, LIEM M, JONG-RAADSEN S A, et al. Rapid de novo assembly of the European eel genome from nanopore sequencing reads [J]. Scientific Reports, 2017, 7: 7213. doi: 10.1038/s41598-017-07650-6 [12] FELLERS J P, WEBB C, FELLERS M C, et al. Wheat virus identification within infected tissue using nanopore sequencing technology [J]. Plant Disease, 2019, 103(9): 2199−2203. doi: 10.1094/PDIS-09-18-1700-RE [13] GIORDANO F, AIGRAIN L, QUAIL M A, et al. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms [J]. Scientific Reports, 2017, 7: 3935. doi: 10.1038/s41598-017-03996-z [14] XIA E H, ZHANG H B, SHENG J, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis [J]. Molecular Plant, 2017, 10(6): 866−877. doi: 10.1016/j.molp.2017.04.002 [15] XIA E H, TONG W, HOU Y, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation [J]. Molecular Plant, 2020, 13(7): 1013−1026. doi: 10.1016/j.molp.2020.04.010 [16] CHEN J D, ZHENG C, MA J Q, et al. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant [J]. Horticulture Research, 2020, 7: 63. doi: 10.1038/s41438-020-0288-2 [17] ZHANG Q J, LI W, LI K, et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution [J]. Molecular Plant, 2020, 13(7): 935−938. doi: 10.1016/j.molp.2020.04.009 [18] ZHANG W Y, ZHANG Y J, QIU H J, et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties [J]. Nature Communications, 2020, 11: 3719. doi: 10.1038/s41467-020-17498-6 [19] GOODWIN S, MCPHERSON J D, MCCOMBIE W R. Coming of age: Ten years of next-generation sequencing technologies [J]. Nature Reviews Genetics, 2016, 17(6): 333−351. doi: 10.1038/nrg.2016.49 [20] RHOADS A, AU K F. PacBio sequencing and its applications [J]. Genomics, Proteomics & Bioinformatics, 2015, 13(5): 278−289. [21] QIAO D H, YANG C, CHEN J, et al. Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant (Camellia sinensis) [J]. Scientific Reports, 2019, 9: 2709. doi: 10.1038/s41598-019-39286-z [22] DHAR R, SEETHY A, PETHUSAMY K, et al. De novo assembly of the Indian blue peacock (Pavo cristatus) genome using Oxford Nanopore technology and Illumina sequencing [J]. GigaScience, 2019, 8(5): giz038. doi: 10.1093/gigascience/giz038 [23] DEBLADIS E, LLAURO C, CARPENTIER M C, et al. Detection of active transposable elements in Arabidopsis thaliana using Oxford Nanopore Sequencing technology [J]. BMC Genomics, 2017, 18(1): 1−8. doi: 10.1186/s12864-016-3406-7 [24] LIAO Y C, LIN S H, LIN H H. Completing bacterial genome assemblies: Strategy and performance comparisons [J]. Scientific Reports, 2015, 5: 8747. doi: 10.1038/srep08747 [25] SHIN S C, AHN D H, KIM S J, et al. Advantages of single-molecule real-time sequencing in high-GC content genomes [J]. PLoS One, 2013, 8(7): e68824. doi: 10.1371/journal.pone.0068824 [26] 庞丹丹, 刘玉飞, 孙云南, 等. 苦茶全长转录组测序及基因结构分析 [J]. 西北农业学报, 2021, 30(4):563−571. doi: 10.7606/j.issn.1004-1389.2021.04.011PANG D D, LIU Y F, SUN Y N, et al. Full-length transcriptome and gene structure analysis of kucha (Camellia sinensis) [J]. Acta Agriculturae Boreali-Occidentalis Sinica, 2021, 30(4): 563−571.(in Chinese) doi: 10.7606/j.issn.1004-1389.2021.04.011 [27] WEI C L, YANG H, WANG S B, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality [J]. PNAS, 2018, 115(18): E4151−E4158. doi: 10.1073/pnas.1719622115 [28] ZHOU X B, LINDSAY H, ROBINSON M D. Robustly detecting differential expression in RNA sequencing data using observation weights [J]. Nucleic Acids Research, 2014, 42(11): e91. doi: 10.1093/nar/gku310 [29] LI H. Minimap2: pairwise alignment for nucleotide sequences [J]. Bioinformatics, 2018, 34(18): 3094−3100. doi: 10.1093/bioinformatics/bty191 [30] THIEL T, MICHALEK W, VARSHNEY R, et al. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) [J]. Theoretical and Applied Genetics, 2003, 106(3): 411−422. doi: 10.1007/s00122-002-1031-0 [31] 陈春林, 田易萍, 陈林波, 等. 基于荧光标记的紫娟茶树转录组EST-SSR标记开发 [J]. 江苏农业学报, 2018, 34(4):747−753. doi: 10.3969/j.issn.1000-4440.2018.04.005CHEN C L, TIAN Y P, CHEN L B, et al. EST-SSR marker development of Zijuan tea tree transcriptome based on the fluorescent labeling [J]. Jiangsu Journal of Agricultural Sciences, 2018, 34(4): 747−753.(in Chinese) doi: 10.3969/j.issn.1000-4440.2018.04.005 [32] 许明, 曾莉, 黄昕颖, 等. 藤茶转录组SSR分子标记开发与初步验证 [J]. 分子植物育种, 2020, 18(19):6441−6448.XU M, ZENG L, HUANG X Y, et al. Development and identification of SSR molecular markers based on transcriptome sequences of Ampelopsis grossedentata [J]. Molecular Plant Breeding, 2020, 18(19): 6441−6448.(in Chinese) [33] 辛静, 李斌, 叶鹏, 等. 云南金花茶转录组序列分析及功能注释 [J]. 经济林研究, 2020, 38(3):85−94.XIN J, LI B, YE P, et al. Transcriptome sequence analysis and functional annotation of Camellia fascicularis H. T. Chang [J]. Non-Wood Forest Research, 2020, 38(3): 85−94.(in Chinese) [34] ZHENG Y, JIAO C, SUN H H, et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases [J]. Molecular Plant, 2016, 9(12): 1667−1670. doi: 10.1016/j.molp.2016.09.014 [35] KONG L, ZHANG Y, YE Z Q, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine [J]. Nucleic Acids Research, 2007, 35(S2): W345−W349. doi: 10.1093/nar/gkm391 [36] SUN L, LUO H T, BU D C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts [J]. Nucleic Acids Research, 2013, 41(17): e166. doi: 10.1093/nar/gkt646 [37] WANG L G, PARK H J, DASARI S, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model [J]. Nucleic Acids Research, 2013, 41(6): e74. doi: 10.1093/nar/gkt006 [38] EL-GEBALI S, MISTRY J, BATEMAN A, et al. The Pfam protein families database in 2019 [J]. Nucleic Acids Research, 2019, 47(D1): D427−D432. doi: 10.1093/nar/gky995 [39] ANDERS S, HUBER W. Differential expression analysis for sequence count data [J]. Nature Precedings, 2010: 1. [40] KASIANOWICZ J J, BEZRUKOV S M. On ‘three decades of nanopore sequencing’ [J]. Nature Biotechnology, 2016, 34(5): 481−482. doi: 10.1038/nbt.3570 [41] MAGI A, SEMERARO R, MINGRINO A, et al. Nanopore sequencing data analysis: State of the art, applications and challenges [J]. Briefings in Bioinformatics, 2018, 19(6): 1256−1272. [42] JAIN M, OLSEN H E, PATEN B, et al. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community [J]. Genome Biology, 2016, 17(1): 1−11. doi: 10.1186/s13059-015-0866-z [43] 朱兴正, 夏丽飞, 陈林波, 等. 保护品种云茶1号茶树全长转录组测序分析 [J]. 茶叶科学, 2018, 38(2):193−201. doi: 10.3969/j.issn.1000-369X.2018.02.011ZHU X Z, XIA L F, CHEN L B, et al. Full-length transcriptome analysis of protected cultivation ‘yuncha 1'(Camellia sinensis var assamica) [J]. Journal of Tea Science, 2018, 38(2): 193−201.(in Chinese) doi: 10.3969/j.issn.1000-369X.2018.02.011 [44] 鞠烨, 江建平, 尹增芳, 等. 孝顺竹笋箨全长转录组测序分析 [J]. 南京林业大学学报(自然科学版), 2020, 44(6):175−183.JU Y, JIANG J P, YIN Z F, et al. Full-length transcriptome sequencing and annotation analyses of Bambusa multiplex sheath [J]. Journal of Nanjing Forestry University (Natural Sciences Edition), 2020, 44(6): 175−183.(in Chinese) [45] 潘敏, 于旭东, 蔡泽坪, 等. 波罗蜜茎叶全长转录组分析 [J]. 热带作物学报, 2020, 41(7):1288−1297. doi: 10.3969/j.issn.1000-2561.2020.07.002PAN M, YU X D, CAI Z P, et al. Transcriptome Data Analysis of Artocarpus heterophyllus stem and leaves [J]. Chines Journal of Tropical Crops, 2020, 41(7): 1288−1297.(in Chinese) doi: 10.3969/j.issn.1000-2561.2020.07.002 -







 下载:
下载: