Identification and Functional Analysis of Genes Related to Cocoon Shell Ratio in Bombyx mori
-
摘要:
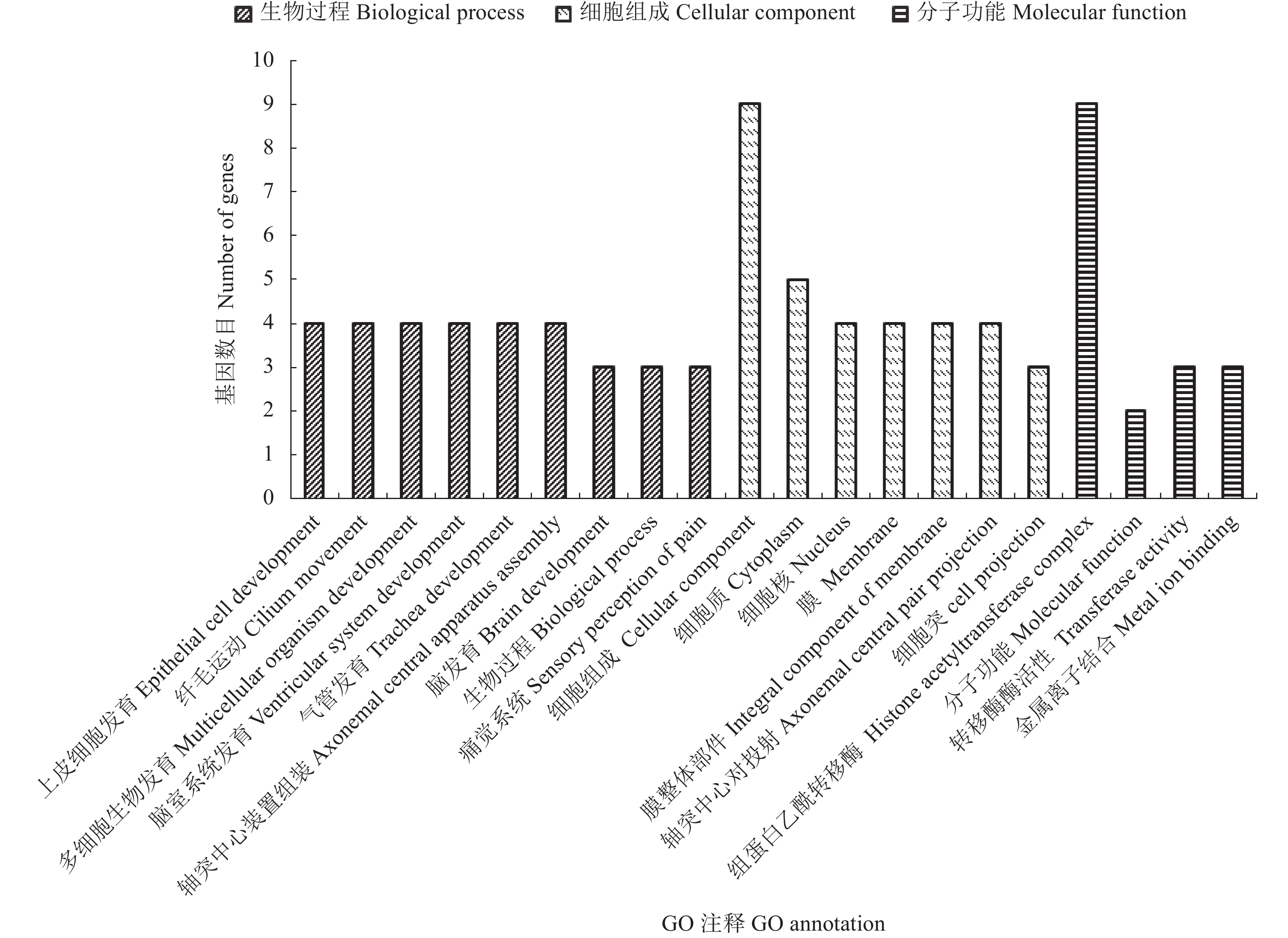
目的 挖掘家蚕茧层率相关基因,以期为家蚕茧层率性状分子遗传改良提供参考依据。 方法 以多丝量家蚕品种菁松和中丝量家蚕品种芙蓉为亲本构建BC1代分离群体。在BC1代雄性群体中挑选极端高/低茧层率个体构建子代DNA混池,运用BSA-seq方法对茧层率关联区域进行定位,并运用BLAST软件对关联区域的编码基因进行GO和KEGG等数据库注释及功能预测。 结果 重测序数据与家蚕参考基因组平均比对率为98.86%,平均基因组覆盖度为95.79%(1×)和88.63%(5×);变异检测共获得26 557 646个SNPs;∆(SNP-index)定位到3个与茧层率显著相关的区域,分别为Chr.2:4430~4930 kb、Chr.4:12350~12920 kb和Chr.13:3230~3730 kb,共包含70个编码基因。通过GO与KEGG注释,有58个基因注释到GO数据库,涉及生物过程、分子功能和细胞组分三大类;有19个基因注释到KEGG通路,分布于34个代谢通路中。通过KEGG代谢通路分析,筛选出10个可能对家蚕茧层率有重要调控作用的基因,推测其参与了家蚕丝腺细胞运动、能量代谢和蛋白质合成加工。 结论 运用BSA-seq方法在家蚕第2、4和13号染色体上定位到与茧层率关联的区域,筛选到10个可能与茧层率密切相关的候选基因,为茧层率关键调控基因精细定位及克隆奠定基础。 Abstract:Objective Genes related to the cocoon shell ratio of Bombyx mori were investigated for improvement on silkworm productivity. Method Segregated BC1 populations of a highly productive silkworm, Jingsong, and a moderately productive Furong were established. DNA pools were constructed by mixing 30 high and low cocoon shell ratio individuals from the BC1 male populations. BSA-seq was applied to identify the regions relevant to the target trait. The coding genes at the candidate regions were annotated with BLAST software in GO and KEGG databases. Result The percentage of clean reads that matched the reference genome of P50 was 98.86%, and those of the average genome coverage 95.79% for 1× and 88.63% for 5×. Variant calling resulted in 26 557 646 SNPs. Three QTLs for the cocoon shell ratio detected by ∆(SNP-index) were in the intervals of 4 430–4930 kb on Chr.2, 12350–12920 kb on Chr.4, and 3230–3730 kb on Chr.13. There were 70 coding genes found in the associated regions. According to the GO database annotation, 58 genes were categorized in the groups of biological process, molecular function, or cellular component. The 19 genes annotated in the KEGG database distributed in 34 metabolic pathways. Of which, 10 might play important roles in regulating the cocoon shell ratio and the functions involving the silk gland cell movement, energy metabolism, and/or protein synthesis. Conclusion The genes related to the silkworm cocoon shell ratio were identified by BSA-seq method. Ten genes were speculated to possibly associate with the silk formation. The present study only managed to locate the regions on chromosomes 2, 4, and 13 of the genes in B. mori that might related to the cocoon shell ratio. Further study will be needed to finely map and clone the key regulatory genes in the highly complex regulatory mechanism. -
Key words:
- Bombyx mori /
- silkworm cocoon shell ratio /
- BSA-seq /
- gene annotation
-
图 1 分子标记及关联信号在染色体上的分布
从外到内依次为参考基因组染色体坐标、染色体上基因分布(颜色越深表示基因密度越大)、SNP密度分布(圆点越密集表示SNP密度越大)、InDel密度分布(三角形越密集表示InDel密度越大)、Index值在染色体上的分布。
Figure 1. Distribution of SNPs, InDels, and associated signals on chromosome
Shown from outside inward: chromosome coordinates of reference genome, genes distribution on chromosome (darker color indicates greater gene density), SNP density distribution (density of dots corresponds to that of SNP), InDel density distribution (density of triangles reflects that of InDel), and distribution of indices on chromosome.
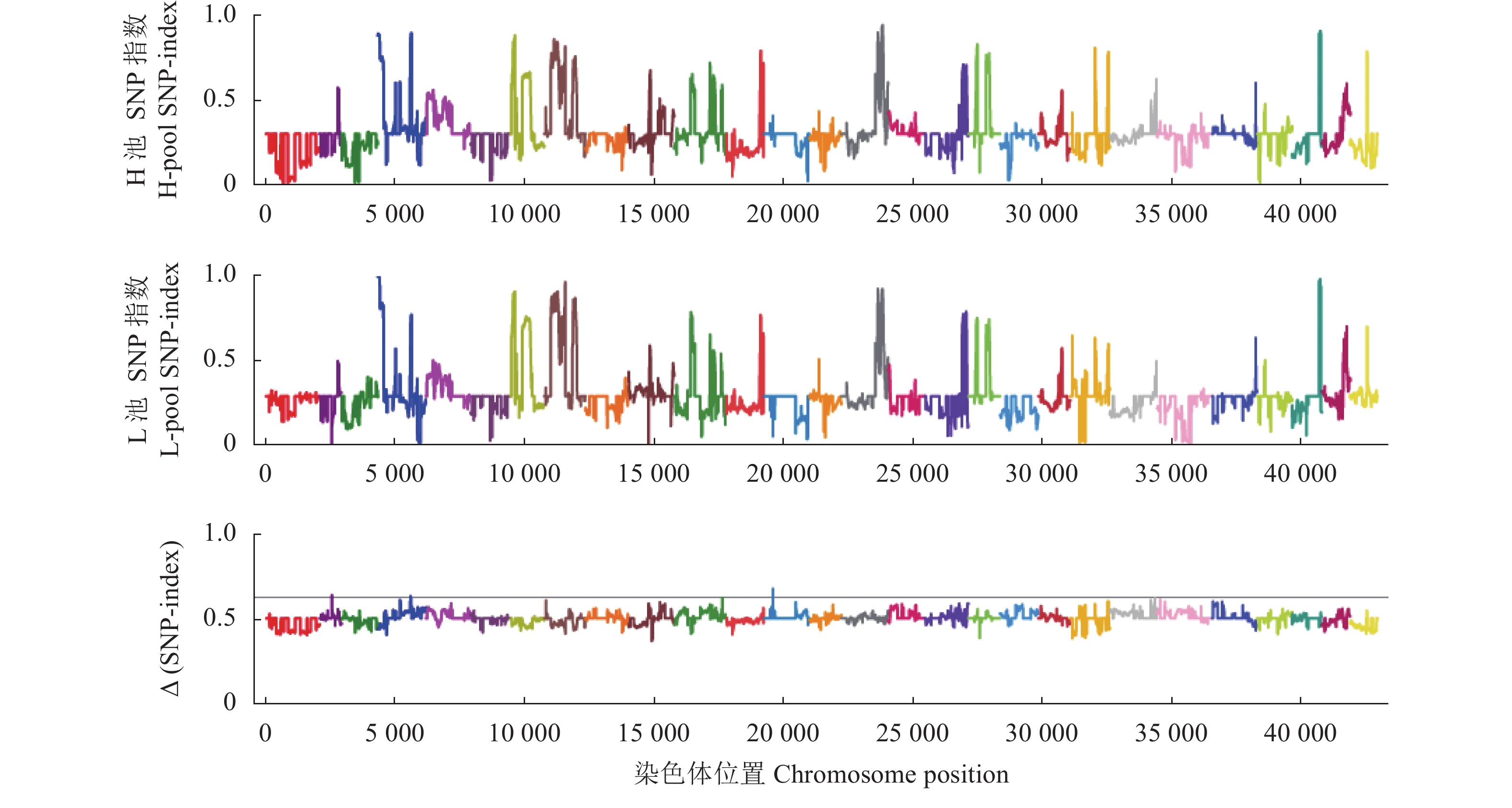
图 2 H池和L池SNP-index、∆(SNP-index)分布情况
图中不同颜色表示不同的染色体,横坐标为1~28号染色体上每个window的具体物理位置,纵坐标为位置所对应的Index值。
Figure 2. SNP-index and ∆(SNP-index) distribution of H-pool and L-pool.
Different colors represent different chromosomes; x-axis is for physical location of each window on chromosomes 1 to 28; y-axis is for Index corresponding to respective locations.
表 1 测序数据质量
Table 1. Statistics on quality of sequencing data
样品编号
Sample ID原始数据量
Raw data/bp过滤后数据量
Clean base/bp原测序reads 数
Raw reads过滤后reads 数
Clean readsQ20/% Q30/% GC含量
GC content/%菁松(JS) 380002170 6295123605 42251670 41738170 98.10 94.04 38.83 芙蓉(FR) 5853501108 5771738817 38764908 38269934 97.91 93.56 38.96 H-pool 16080222540 15911340150 106491540 105500096 98.13 94.11 38.89 L-pool 16401318000 16232070628 108618000 107624014 98.13 94.11 38.66 Q20:高质量测序数据中质量值≥20的碱基所占百分比;Q30:高质量测序数据中质量值≥30的碱基所占百分比。
Q20:The percentage of the bases whose Phred value are more than 20; Q30:The percentage of the bases whose Phred value are more than 30.表 2 质控数据与参考基因组比对情况
Table 2. Matching between quality control data and reference genome
样品编号
Sample ID比对率
Mapped ratio/%比对到基因组上的reads比例
Properly ratio/%平均测序深度
Average depth基因组覆盖度(1×)
Genome coverage(1×) /%基因组覆盖度(5×)
Genome coverage(5×) /%菁松(JS) 98.84 82.72 10.34 94.70 84.71 芙蓉(FR) 98.88 82.28 9.44 94.41 82.61 H-pool 98.90 81.29 26.02 97.04 93.59 L-pool 98.82 82.38 27.59 97.02 93.59 1×覆盖度:1 个碱基覆盖的位点占基因组的百分比;5×覆盖度:5个碱基覆盖的位点占基因组的百分比。
Coverage 1×: the percentage of at least 1 base-covered site in reference genome; Coverage 5×: the percentage of at least 5 base-covered sites in reference genome.表 3 关联区域信息统计
Table 3. Statistics of the related genes
染色体编号
Chromosome ID关联区域起点
Start of associated
regions/bp关联区域终点
End of associated
regions/bp关联区域长度
Associated region
size/MbSNP数量
SNP number关联区域内基因个数
Gene number in the
associated regions第13染色体 Chr 13 3230000 3730000 0.50 11 13 第4染色体 Chr 4 12350000 12920000 0.50 39 48 第2染色体 Chr 2 4430000 4930000 0.57 17 9 合计 Total 1.57 67 70 表 4 候选基因的KEGG通路分析
Table 4. KEGG pathway of genes in candidate regions
一级代谢
Primary metabolism二级代谢
Secondary metabolism三级代谢
Tertiary metabolism通路编号
Ko ID基因编号
Gene ID新陈代谢
Metabolism聚糖生物合成与代谢
Glycan biosynthesis and metabolism糖胺聚糖降解
Glycosaminoglycan degradationko00531 KWMTBOMO07657 O-聚糖生物合成
Other types of O-glycan biosynthesisko00514 KWMTBOMO02149 脂质代谢
Lipid metabolism初级胆汁酸生物合成
Primary bile acid biosynthesisko00120 KWMTBOMO02138 有机体系统
Organismal systems免疫系统
Immune systemRIG-I样受体信号通路
RIG-I-like receptor signaling pathwayKo04622 KWMTBOMO02145;
KWMTBOMO07659;
KWMTBOMO02143Toll样受体信号通路
Toll-like receptor signaling pathwayko04620 KWMTBOMO07659 NOD样受体信号通路
NOD-like receptor signaling pathwayko04621 KWMTBOMO07659 内分泌系统
Endocrine system胰高血糖素信号通路
Glucagon signaling pathwayKo04922 KWMTBOMO07653;
KWMTBOMO07652;
KWMTBOMO02146甲状腺激素信号通路
Thyroid hormone signaling pathwayko04919 KWMTBOMO07653;
KWMTBOMO07652胰岛素信号通路
Insulin signaling pathwayko04910 KWMTBOMO02146 神经系统
Nervous system长时程增强效应
Long-term potentiationko04720 KWMTBOMO07653;
KWMTBOMO07652神经营养因子信号通路
Neurotrophin signaling pathwayko04722 KWMTBOMO07659 环境适应
Environmental adaptation生理节律 Circadian rhythm ko04710 KWMTBOMO00853 环境信息加工
Environment information processing信号转导
Signal transductionRas信号通路 Ras signaling pathway ko04014 KWMTBOMO02152 丝裂原活化蛋白激酶信号通路
MAPK signaling pathwayKo04013 KWMTBOMO02145;
KWMTBOMO07659;
KWMTBOMO02143钙离子信号通路
Calcium signaling pathwayko04020 KWMTBOMO02146 低氧诱导因子-1信号通路
HIF-1 signaling pathwayko04066 KWMTBOMO07653;
KWMTBOMO07652Wnt信号通路
Wnt signaling pathwayko04310 KWMTBOMO07653;
KWMTBOMO07652;
KWMTBOMO00853环磷酸腺苷信号通路
cAMP signaling pathwayko04024 KWMTBOMO07653;
KWMTBOMO07652Notch信号通路 Notch signaling pathway ko04330 KWMTBOMO07653;
KWMTBOMO07652Jak-STAT信号通路
Jak-STAT signaling pathwayko04630 KWMTBOMO07653;
KWMTBOMO07652TGF-β信号通路
TGF-beta signaling pathwayko04350 KWMTBOMO07653;
KWMTBOMO00853;
KWMTBOMO07652刺猬信号通路
Hedgehog signaling pathwayko04341 KWMTBOMO00853 细胞进程
Cellular processes细胞生长和死亡
Cell growth and death细胞周期 Cell cycle Ko04110 KWMTBOMO07653;
KWMTBOMO07652;
KWMTBOMO00853细胞凋亡 Apoptosis ko04214 KWMTBOMO07659 卵母细胞减数分裂 Oocyte meiosis ko04114 KWMTBOMO00853 细胞通讯 Cell communication 黏着连接 Adherens junction ko04520 KWMTBOMO07653;
KWMTBOMO07652运输与分解代谢
Transport and catabolism内吞作用 Endocytosis ko04144 KWMTBOMO07659 溶酶体 Lysosome ko04142 KWMTBOMO07657 遗传信息加工 Genetic information processing 折叠、组装和降解
Folding, sorting and degradation泛素介导的蛋白质水解
Ubiquitin mediated proteolysisko04120 KWMTBOMO07659;
KWMTBOMO02114;
KWMTBOMO00853蛋白质在内质网上的加工
Protein processing in endoplasmic reticulumko04141 KWMTBOMO02147;
KWMTBOMO00853蛋白酶体 Proteasome ko03050 KWMTBOMO02148 转录 Transcription 转录因子 Basal transcription factors ko03022 KWMTBOMO02120 剪接体 Spliceosome ko03040 KWMTBOMO02133 翻译 Translation 核糖体 Ribosome ko03010 KWMTBOMO02140 -
[1] 任晓晓, 罗朝斌, 孙运朋, 等. 高原蚕区家蚕茧层率遗传分析 [J]. 农学学报, 2020, 10(6):75−80.REN X X, LUO C B, SUN Y P, et al. The cocoon shell ratio of Bombyx mori from sericultural area of plateau: Genetic analysis [J]. Journal of Agriculture, 2020, 10(6): 75−80.(in Chinese) [2] LI C L, TONG X L, ZUO W D, et al. QTL analysis of cocoon shell weight identifies BmRPL18 associated with silk protein synthesis in silkworm by pooling sequencing [J]. Scientific Reports, 2017, 7: 17985. doi: 10.1038/s41598-017-18277-y [3] 刘娜, 李娟, 秦笙, 等. 家蚕茧丝相关性状的研究进展 [J]. 中国蚕业, 2016, 37(4):6−9. doi: 10.16839/j.cnki.zgcy.2016.04.002LIU N, LI J, QIN S, et al. Research progress on cocoon silk related traits of silkworm [J]. China Sericulture, 2016, 37(4): 6−9.(in Chinese) doi: 10.16839/j.cnki.zgcy.2016.04.002 [4] 栾悦, 李春林, 代方银. 家蚕茧丝性状的遗传基础研究 [J]. 蚕学通讯, 2017, 37(1):21−28. doi: 10.3969/j.issn.1006-0561.2017.01.005LUAN Y, LI C L, DAI F Y. Basic researches of the genetics of cocoon traits in silkworm [J]. Newsletter of Sericultural Science, 2017, 37(1): 21−28.(in Chinese) doi: 10.3969/j.issn.1006-0561.2017.01.005 [5] 鲁成, 李斌, 赵爱春, 等. 家蚕重要经济性状的QTL定位研究 [J]. 中国科学(C辑:生命科学), 2004, 34(3):236−242.LU C, LI B, ZHAO A C, et al. QTL mapping of important economic characters of Bombyx mori [J]. Science in China(SerC), 2004, 34(3): 236−242.(in Chinese) [6] 李斌, 鲁成, 赵爱春, 等. 家蚕全茧量及重要相关经济性状的多重区间作图分析 [J]. 中国农业科学, 2005, 38(7):1474−1479. doi: 10.3321/j.issn:0578-1752.2005.07.030LI B, LU C, ZHAO A C, et al. Multiple interval mapping for whole cocoon weight and related economically important traits QTL in silkworm (Bombyx mori) [J]. Scientia Agricultura Sinica, 2005, 38(7): 1474−1479.(in Chinese) doi: 10.3321/j.issn:0578-1752.2005.07.030 [7] 司马杨虎, 李斌, 徐海明, 等. 家蚕茧质性状的QTL定位研究 [J]. 遗传学报, 2005, 32(6):625−632.SIMA Y H, LI B, XU H M, et al. Study on location of QTLs controlling cocoon traits in silkworm [J]. Acta Genetica Sinica, 2005, 32(6): 625−632.(in Chinese) [8] 侯成香, 王修业, 李冰, 等. 家蚕茧丝相关性状的性连锁QTLs定位与分析 [J]. 蚕业科学, 2013, 39(1):35−39. doi: 10.13441/j.cnki.cykx.2013.01.019HOU C X, WANG X Y, LI B, et al. Mapping and analysis of Bombyx mori sex-linked QTLs related to cocoon and silk traits [J]. Science of Sericulture, 2013, 39(1): 35−39.(in Chinese) doi: 10.13441/j.cnki.cykx.2013.01.019 [9] LI B, WANG X Y, HOU C X, et al. Genetic analysis of quantitative trait loci for cocoon and silk production quantity in Bombyx mori (Lepidoptera: Bombycidae) [J]. European Journal of Entomology, 2013, 110(2): 205−213. doi: 10.14411/eje.2013.031 [10] 张之昊, 王俊, 刘章雄, 等. 基于BSA-Seq技术挖掘大豆中黄622的多小叶基因 [J]. 作物学报, 2020, 46(12):1839−1849.ZHANG Z H, WANG J, LIU Z X, et al. Mapping of an incomplete dominant gene controlling multifoliolate leaf by BSA-Seq in soybean(Glycine max L.) [J]. Acta Agronomica Sinica, 2020, 46(12): 1839−1849.(in Chinese) [11] 贾秀苹, 卯旭辉, 岳云, 等. 利用BSA-Seq方法鉴定向日葵耐盐候选基因 [J]. 中国油料作物学报, 2018, 40(6):777−784. doi: 10.7505/j.issn.1007-9084.2018.06.006JIA X P, MAO X H, YUE Y, et al. Identification of major salt-tolerant genes via BSA-Seq method in sunflower [J]. Chinese Journal of Oil Crop Sciences, 2018, 40(6): 777−784.(in Chinese) doi: 10.7505/j.issn.1007-9084.2018.06.006 [12] 徐剑文, 刘剑光, 赵君, 等. 利用BSA-seq发掘棉花适宜机采的果枝长度相关QTL [J]. 棉花学报, 2019, 31(4):319−326. doi: 10.11963/1002-7807.xjwxsh.20190611XU J W, LIU J G, ZHAO J, et al. The identification of QTL associated with cotton fruit branch length suitable for mechanized harvest utilizing BSA-seq [J]. Cotton Science, 2019, 31(4): 319−326.(in Chinese) doi: 10.11963/1002-7807.xjwxsh.20190611 [13] YANG T T, AMANULLAH S, PAN J H, et al. Identification of putative genetic regions for watermelon rind hardness and related traits by BSA-seq and QTL mapping [J]. Euphytica, 2021, 217(2): 19. doi: 10.1007/s10681-020-02758-9 [14] 刘梦雨, 刘小丰, 江东, 等. 利用重测序-BSA分析鉴定金柑油胞发育相关基因 [J]. 园艺学报, 2019, 46(5):841−854.LIU M Y, LIU X F, JIANG D, et al. Identification of genes related to oil gland development in kumquat by using BSA-seq [J]. Acta Horticulturae Sinica, 2019, 46(5): 841−854.(in Chinese) [15] 许芸梅, 李玉梅, 贾玉鑫, 等. 马铃薯红色薯肉调控基因的精细定位与候选基因分析 [J]. 中国农业科学, 2019, 52(15):2678−2685. doi: 10.3864/j.issn.0578-1752.2019.15.011XU Y M, LI Y M, JIA Y X, et al. Fine mapping and candidate genes analysis for regulatory gene of anthocyanin synthesis in red-colored Tuber flesh [J]. Scientia Agricultura Sinica, 2019, 52(15): 2678−2685.(in Chinese) doi: 10.3864/j.issn.0578-1752.2019.15.011 [16] 柳海东, 赵绪涛, 杜德志. 利用QTL-seq技术定位甘蓝型春油菜早花位点cqDTFC8及其近等基因系构建 [J]. 植物生理学报, 2020, 56(2):219−234. doi: 10.13592/j.cnki.ppj.2019.0398LIU H D, ZHAO X T, DU D Z. Mapping of early flowering site cqDTFC8 using QTL-seq technique and construction of its near-isogenic lines in Brassica napus [J]. Plant Physiology Journal, 2020, 56(2): 219−234.(in Chinese) doi: 10.13592/j.cnki.ppj.2019.0398 [17] 尹明智, 胡燕. 基于BSA-seq法的油菜野芥胞质雄性不育恢复基因的分析 [J]. 西北植物学报, 2020, 40(7):1148−1156.YIN M Z, HU Y. Location analysis of restorer gene of Sinapis arvensis cytoplasmic male sterility in Brassica napus based on BSA-seq method [J]. Acta Botanica Boreali-Occidentalia Sinica, 2020, 40(7): 1148−1156.(in Chinese) [18] 欧点点, 赵光伟, 贺玉花, 等. 甜瓜果皮颜色遗传分析及基因定位 [J]. 中国农学通报, 2019, 35(13):64−69. doi: 10.11924/j.issn.1000-6850.casb18110127OU D D, ZHAO G W, HE Y H, et al. Genetic analysis and gene mapping for melon rind color [J]. Chinese Agricultural Science Bulletin, 2019, 35(13): 64−69.(in Chinese) doi: 10.11924/j.issn.1000-6850.casb18110127 [19] 祝新荣, 何克荣, 柳新菊, 等. 多丝量雄蚕新品种华菁×平72的育成 [J]. 蚕业科学, 2014, 40(2):248−253. doi: 10.13441/j.cnki.cykx.2014.02.012ZHU X R, HE K R, LIU X J, et al. Breeding of new male silkworm variety Huajing × Ping 72 with high silk yield [J]. Science of Sericulture, 2014, 40(2): 248−253.(in Chinese) doi: 10.13441/j.cnki.cykx.2014.02.012 [20] 司马杨虎, 徐海明, 赵爱春, 等. 性别效应对家蚕茧质性状QTL定位的影响 [J]. 蚕业科学, 2009, 35(4):783−789. doi: 10.3969/j.issn.0257-4799.2009.04.012SIMA Y H, XU H M, ZHAO A C, et al. Influence of sex-effects on QTL mapping of silkworm cocoon quality traits [J]. Science of Sericulture, 2009, 35(4): 783−789.(in Chinese) doi: 10.3969/j.issn.0257-4799.2009.04.012 [21] ZHAN S, HUANG J H, GUO Q H, et al. An integrated genetic linkage map for silkworms with three parental combinations and its application to the mapping of single genes and QTL [J]. BMC Genomics, 2009, 10: 389. doi: 10.1186/1471-2164-10-389 [22] FANG S M, ZHOU Q Z, YU Q Y, et al. Genetic and genomic analysis for cocoon yield traits in silkworm [J]. Scientific Reports, 2020, 10: 5682. doi: 10.1038/s41598-020-62507-9 [23] 马倩, 马俐, 李胜, 等. 基于RNA-Seq分析Ras1 CA在家蚕后部丝腺过表达对细胞周期通路基因的影响 [J]. 应用昆虫学报, 2015, 52(2):390−399. doi: 10.7679/j.issn.2095-1353.2015.043MA Q, MA L, LI S, et al. RNA-Seq technology based transcriptomic analysis of differentially expressed genes in the cell cycle pathway of Ras1 CA-overexpressed and wild type posterior silk glands of Bombyx mori [J]. Chinese Journal of Applied Entomology, 2015, 52(2): 390−399.(in Chinese) doi: 10.7679/j.issn.2095-1353.2015.043 [24] 张祥乐, 马俐, 马倩, 等. Ras信号通路通过激活转录因子Myc促进核内复制细胞生长 [J]. 昆虫学报, 2018, 61(8):885−894.ZHANG X L, MA L, MA Q, et al. Ras signaling pathway promotes the growth of endoreplication cells through activating the expression of transcription factor Myc [J]. Acta Entomologica Sinica, 2018, 61(8): 885−894.(in Chinese) [25] CALDWELL P E, WALKIEWICZ M, STERN M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release [J]. Current Biology, 2005, 15(20): 1785−1795. doi: 10.1016/j.cub.2005.09.011 [26] MA L, XU H F, ZHU J Q, et al. Ras1 CA overexpression in the posterior silk gland improves silk yield [J]. Cell Research, 2011, 21(6): 934−943. doi: 10.1038/cr.2011.36 -








 下载:
下载: