Transcriptome-based Identification and Expressions of Amaranth TCP Gene Family
-
摘要:
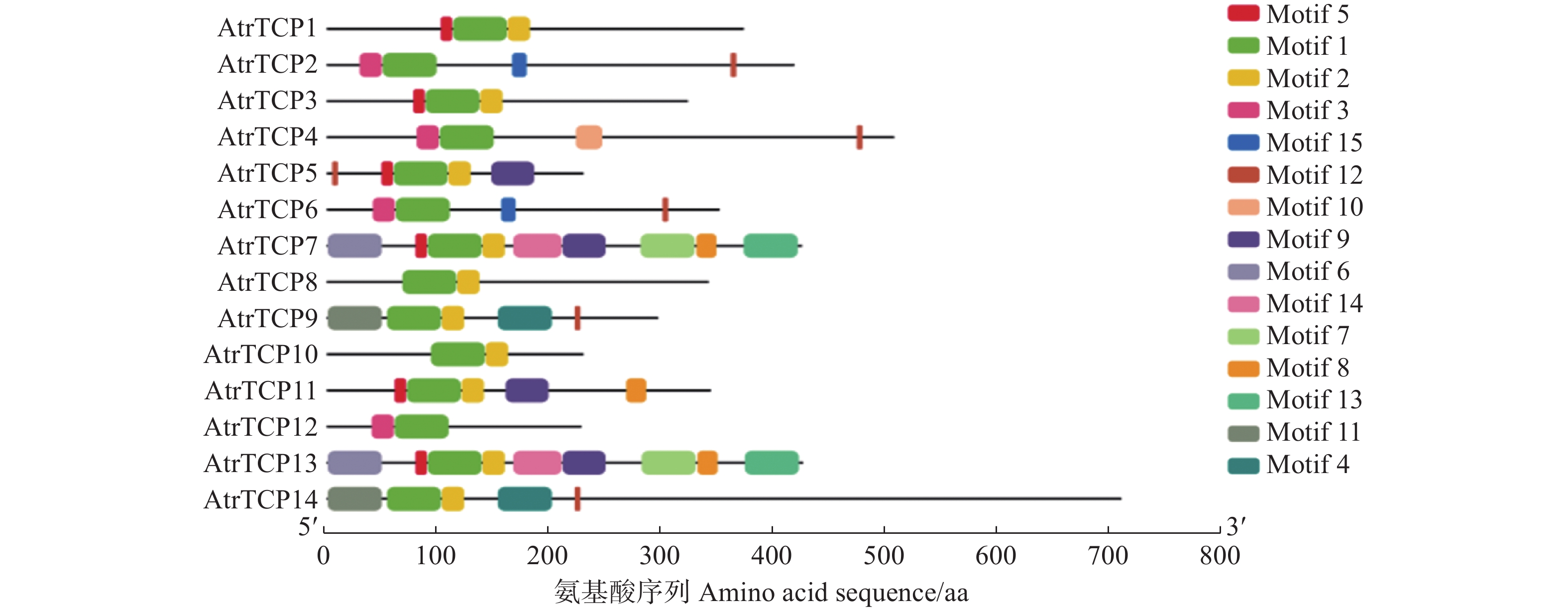
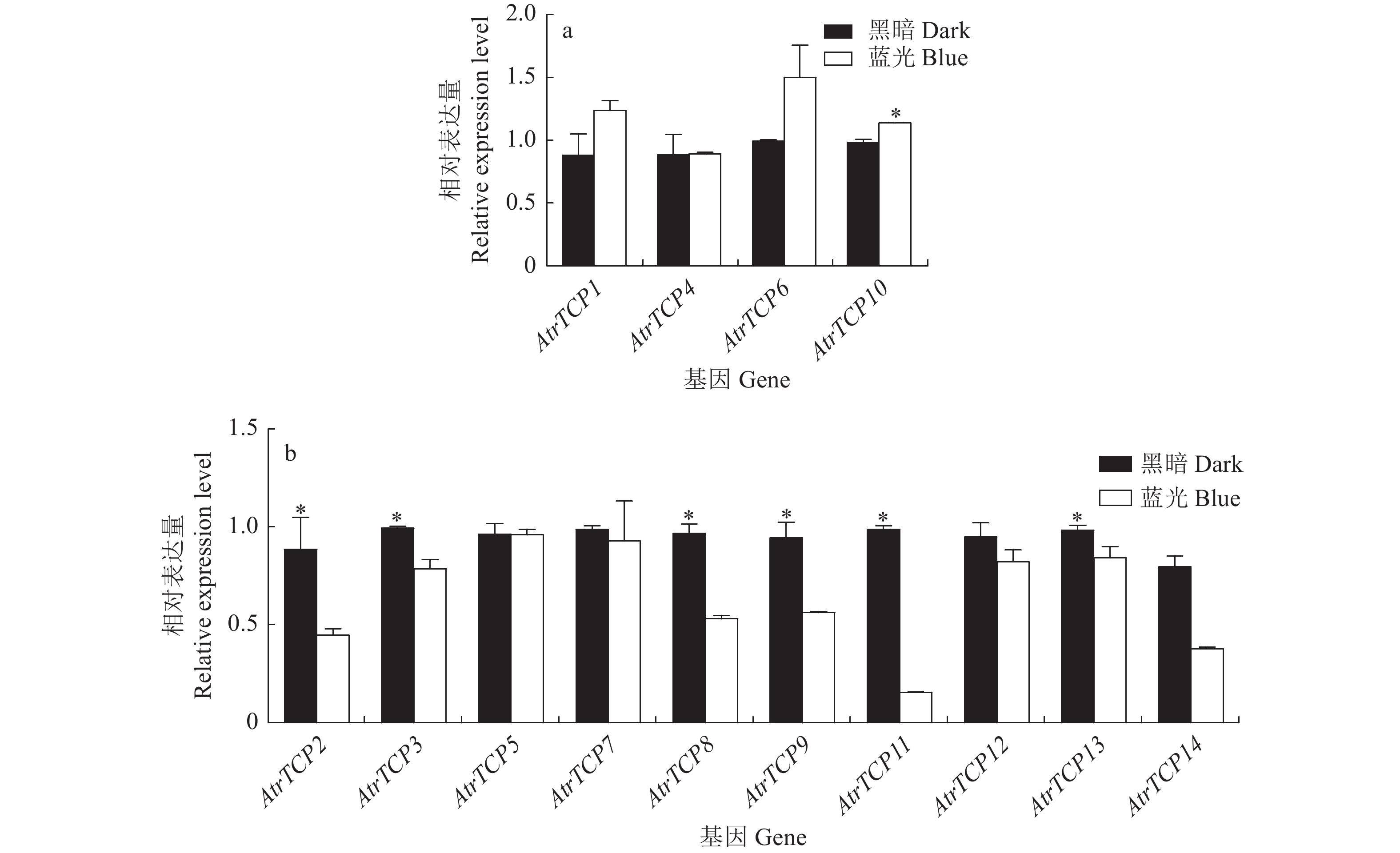
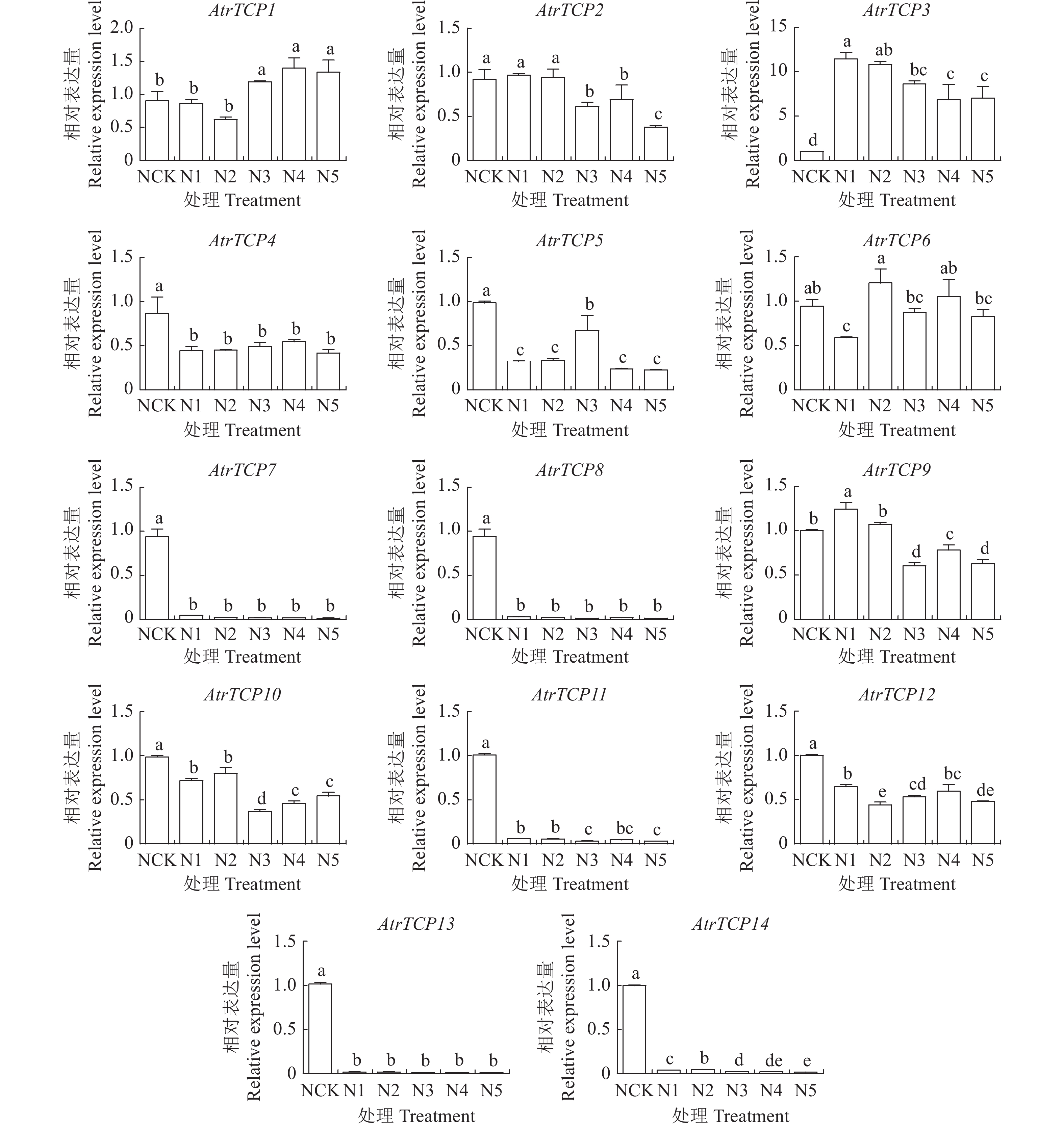
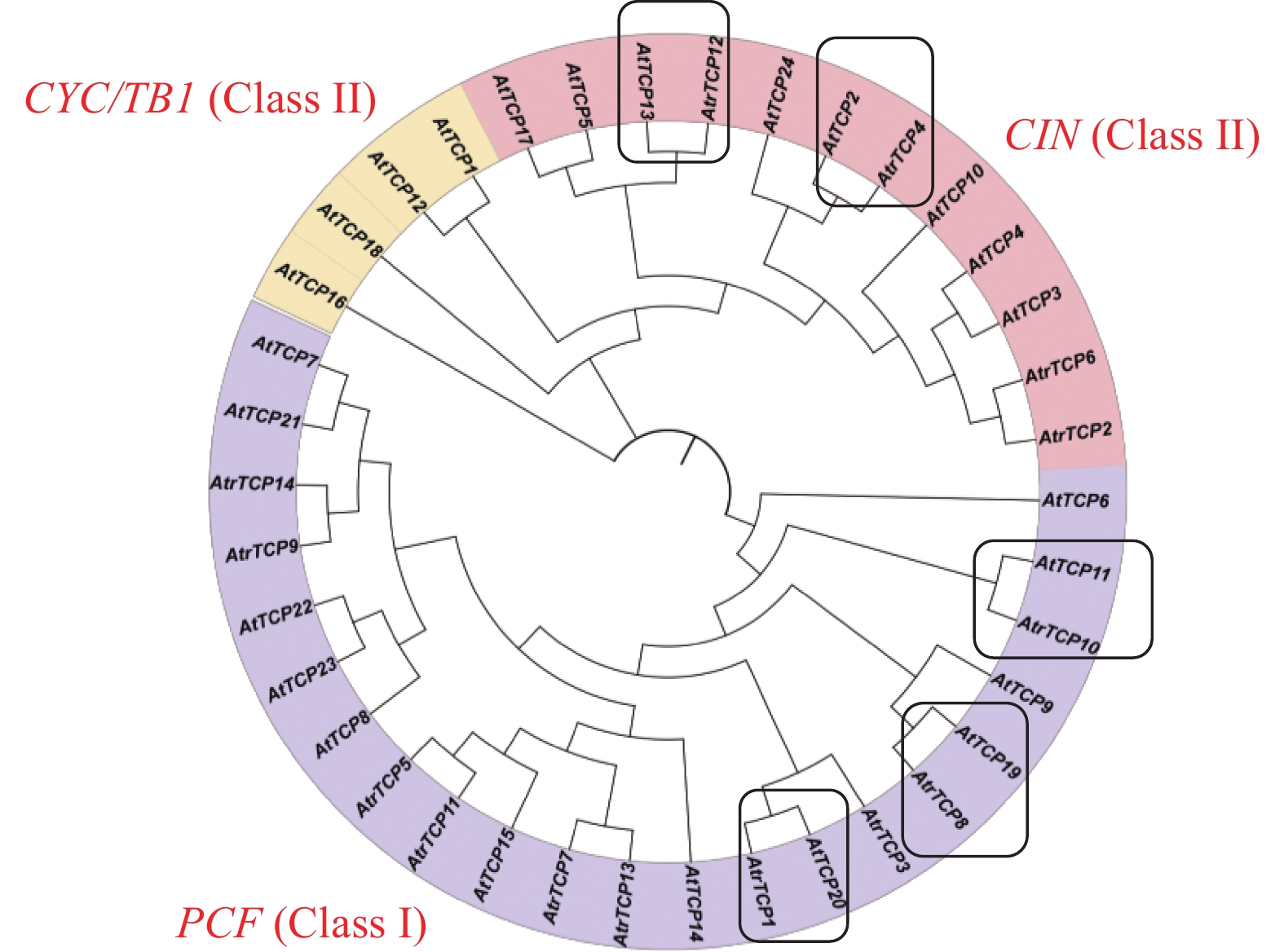
目的 基于大红苋菜转录组数据库,对苋菜TCP基因家族成员进行筛选鉴定,为深入探讨苋菜TCP基因家族对苋菜生长发育及非生物胁迫响应提供参考。 方法 在转录组数据库中,筛选鉴定TCP基因家族成员,并对其进行生物信息学分析;在不同条件处理下培养苋菜幼苗,并利用qRT-PCR技术分析其在不同处理下的表达模式。 结果 苋菜TCP基因家族有14个成员,全部定位于细胞核;AtrTCP蛋白质长度在230~721 aa区间内,预测相对分子量为25.25~78.57 kD,等电点为6.15~9.45,包含15个保守基序;除AtrTCP11为稳定蛋白外,其余均为不稳定蛋白;miRNA预测到两个AtrTCP家族成员AtrTCP2和AtrTCP6为miR319靶基因。qRT-PCR结果显示:在盐溶液不同浓度(0、50、100 、200 mmol·L-1)处理下,该基因家族具有不同的表达模式;在蓝光处理下,AtrTCP10基因表达量上调达到了显著水平;AtrTCP2、AtrTCP3、AtrTCP8、AtrTCP9、AtrTCP11和AtrTCP13表达量下调达到了显著水平;在不同铵硝比(0∶0、0∶10、3∶7、5∶5、7∶3、10∶0)处理下,TCP基因家族各成员表达模式不同,其中,AtrTCP3参与整个氮代谢过程;在苋菜各部位进行qRT-PCR分析发现:AtrTCP3、AtrTCP12 在叶片中高表达,其余家族成员均在根部的表达量较高。 结论 在不同条件处理下,苋菜TCP基因家族各成员表达模式不同,表明苋菜TCP基因家族各成员可能广泛参与非生物胁迫。 Abstract:Objective Members of the amaranth TCP gene family were identified based on the transcriptome database for an in-depth understanding on the roles they associated with the growth, development, and abiotic stress of the plant. Method Members of the TCP gene family were identified by screening the transcriptome database on Dahong amaranth (Amaranthus tricolor L.) for a bioinformatics analysis. Amaranth seedlings were cultured under different conditions and their expressions under different treatments analyzed by qRT-PCR. Results Amaranth TCP gene family has 14 members, all localized in the nucleus. The length of AtrTCP protein was in the range of 230–721 aa, the predicted relative molecular weight was 25.25–78.57 kD, the isoelectric point was 6.15–9.45, and it contained 15 conserved motifs. Except AtrTCP11, which was stable protein, all the other proteins were unstable. Two AtrTCP family members, AtrTCP2 and AtrTCP6, were predicted as miR319 target genes by miRNA. qRT-PCR showed differentiated expressions of the family members under salt solutions of varied concentrations (i.e., 0, 50, 100, and 200 mmol·L−1). The expression of the upregulated AtrTCP10 reached a significant level under blue light, and so did those of the downregulated AtrTCP2, AtrTCP3, AtrTCP8, AtrTCP9, AtrTCP11, and AtrTCP13. Under different concentrations of ammonium nitrate ratio (0∶0, 0∶10, 3∶7, 5∶5, 7∶3, 10∶0) treatment, the expressions of the TCP genes differed. AtrTCP3 was shown to be involved in the entire nitrogen metabolism process. In tissues of amaranth, AtrTCP3 and AtrTCP12 were highly expressed in leaves, while the others in roots. Conclusion Depending on the treatments, the expressions of the amaranth TCP gene family members differed. It suggested a broad spectrum of involvements by the members associated with abiotic stresses on amaranth. -
Key words:
- Amaranthus tricolor L. /
- transcription factor /
- gene family /
- bioinformatics analysis /
- expression
-
表 1 AtrTCP定量引物
Table 1. Quantitative primers of AtrTCP
基因
Gene正向引物序列(5′-3′)
Forward primer sequences (5′-3′)反向引物序列(5′-3′)
Reverse primer sequences(5′-3′)AtrTCP1 GCTGCTACTGGTGCTTCTATT ACCCATACCCATACCCATC AtrTCP2 GATGAGCAGCAACATTCACT CACCACTTCCACCAACACT AtrTCP3 TCTTCAACAAGCCGAACC ATTTCCCACCTCAATCCC AtrTCP4 GGTTAGTTCAAACGGCAGA ACTGTCTTCCAAATCGGGT AtrTCP5 TTATCCATACCCGAACAGC TTTAGTGTGACGGTCTTTGG AtrTCP6 TTGCTACTGGTAGAGGTGGT CCGGACCCATGTAACATTGA AtrTCP7 AACCTCCACCAAAGACCGT GTTTCTCCATCTGACTTATGCC AtrTCP8 GCCTACAGAAACAGTAACCCTC CCCGTTCAAGAAGCCATT AtrTCP9 TCCACCAAGGTCATCATCTT ATCATCGTCTCTCCGTCCA AtrTCP10 CGCAGAATCAAGAACAAGC TGACTTTGGTATGGCGGT AtrTCP11 TGCTACAGGAACCGGGACTATT GATGGCTTGGATCACTCAACTG AtrTCP12 TGGAGGCAAAGATAGACATAGC TAATGGGCAAAGGAGGGA AtrTCP13 CAACCATCATCGTCCCAA AGTTCCACTTTGCGGCTGT AtrTCP14 ACCACAGCCACAACAAAGT GCGGATACTGAGTCAAAGGA EF1a GGGATGCTGGTATGGTGAA ACGGGTCATTTCTTCTTCTGAG 表 2 AtrTCP基因家族理化性质分析
Table 2. Physicochemical properties of AtrTCP family
基因 ID
Gene ID基因名称
Gene
name蛋白质长度
Protein
length/aa分子式
Formula理化性质
Physicochemical properties相对分子量
Relative molecular
weight/kD等电点
Theoretical
pI不稳定系数
Instability
index平均亲水系数
Grand average of
hydropathicityCL11166.Contig2_All AtrTCP1 377 C1747H2777N535O570S14 40.84 6.61 50.76 −0.712 CL1181.Contig4_All AtrTCP2 423 C1950H3008N608O636S6 45.34 6.64 54.40 −0.805 CL4511.Contig1_All AtrTCP3 326 C1520H2432N462O495S6 35.29 9.17 50.51 −0.540 CL531.Contig1_All AtrTCP4 514 C2309H3640N736O795S14 54.88 8.57 43.25 −0.823 CL7519.Contig2_All AtrTCP5 231 C1096H1711N341O342S9 25.43 9.00 49.08 −0.764 CL7919.Contig2_All AtrTCP6 355 C1671H2575N519O543S9 38.91 6.79 40.47 −0.860 CL9396.Contig1_All AtrTCP7 430 C1951H3036N616O654S9 45.87 6.66 47.14 −0.747 Unigene1202_All AtrTCP8 345 C1544H2522N462O518S9 36.13 9.30 47.05 −0.408 Unigene22459_All AtrTCP9 299 C1373H2214N422O444S4 31.87 9.45 56.13 −0.563 Unigene27038_All AtrTCP10 231 C1115H1771N317O340S6 25.25 9.23 63.06 −0.492 Unigene27184_All AtrTCP11 347 C1606H2483N485O523S10 37.27 8.49 38.62 −0.743 Unigene35431_All AtrTCP12 230 C1118H1787N329O364S4 25.79 8.56 44.38 −0.820 Unigene6554_All AtrTCP13 431 C1955H3042N618O656S9 45.99 6.66 47.05 −0.754 Unigene73441_All AtrTCP14 721 C3395H5434N1006O1105S17 78.57 6.15 58.04 −0.602 表 3 AtrTCP基因家族蛋白质二级结构及亚细胞定位分析
Table 3. Protein secondary structures and subcellular localizations of AtrTCP family
转录因子
Transcription
factorTCP结构域
TCP domain蛋白质二级结构
Secondary protein structure亚细胞定位
Subcellulav
localizationα-螺旋
Alpha helix/%β-转角
Beta turn/%延伸链
Extended strand/%无规卷曲
Random coil/%AtrTCP1 111-377 19.36 5.04 13.79 61.80 Nucleus. AtrTCP2 43-178 14.42 2.36 11.58 71.63 Nucleus. AtrTCP3 86-326 24.23 5.52 15.34 54.91 Nucleus. AtrTCP4 95-253 14.20 2.53 15.18 68.09 Nucleus. AtrTCP5 57-228 13.85 9.52 16.45 60.17 Nucleus. AtrTCP6 55-223 14.08 1.69 14.65 69.58 Nucleus. AtrTCP7 88-290 20.70 4.19 10.23 64.88 Nucleus. AtrTCP8 65-176 15.65 5.22 10.43 68.70 Nucleus. AtrTCP9 51-258 19.73 6.02 18.06 56.19 Nucleus. AtrTCP10 91-231 19.05 4.76 13.42 62.77 Nucleus. AtrTCP11 69-256 11.82 10.95 21.33 55.91 Nucleus. AtrTCP12 54-215 22.61 5.22 13.48 58.70 Nucleus. AtrTCP13 88-291 14.39 5.10 11.37 69.14 Nucleus. AtrTCP14 51-332 20.11 5.83 16.37 57.70 Nucleus. 表 4 苋菜AtrTCP基因家族miRNA的预测及差异表达分析
Table 4. Prediction and differential expressions of AtrTCP family miRNA
基因名称
Gene name对应
miRNA期望值
Expectation抑制
Inhibition多样性
Multiplicity表达量
Expression quantity表达趋势
Express trendAtrTCP6 miR319_1 1.5 切割 Cleavage 1 0.764797327 上调 Up miR319_2 1.5 切割 Cleavage 1 −0.396055889 下调 Down miR319a-3p 1.5 切割 Cleavage 1 −0.134714289 下调 Down miR319a_1 1.5 切割 Cleavage 1 0.179984679 上调 Up miR319a 2.5 切割 Cleavage 1 −0.06725778 下调 Down miR319c_3 2.5 切割 Cleavage 1 0.137854295 上调 Up miR159a_1 3.0 切割 Cleavage 1 0.073891584 上调 Up Total miRNA — — ― 0.558499927 上调 Up AtrTCP2 miR319_1 3.0 切割 Cleavage 1 0.764797327 上调 Up miR319_2 3.0 切割 Cleavage 1 −0.396055889 下调 Down miR319a-3p 3.0 切割 Cleavage 1 −0.134714289 下调 Down miR319a_1 3.0 切割 Cleavage 1 0.179984679 上调 Up Total miRNA — — — 0.414011828 上调 Up AtrTCP11 miR5658 3.0 切割 Cleavage 1 −0.227576991 下调 Down AtrTCP13 miR5658 3.0 切割 Cleavage 1 −0.227576991 下调 Down “Total miRNA”表示基因对应miRNA的总表达趋势;“—”表示无结果。
"Total miRNA" represents the Total expression trend of miRNA corresponding to the gene. "—" indicates no result. -
[1] 仝亚楠, 刘雪, 刘秀洁, 等. 苋菜原生质体和瞬时转化体系的建立及应用 [J]. 分子植物育种, 2021, 19(19):6476−6481. doi: 10.13271/j.mpb.019.006476TONG Y N, LIU X, LIU X J, et al. Establishment and application of protoplast and transient transformation system of edible amaranth (Amaranthus spp. ) [J]. Molecular Plant Breeding, 2021, 19(19): 6476−6481.(in Chinese) doi: 10.13271/j.mpb.019.006476 [2] STRACK D, VOGT T, SCHLIEMANN W. Recent advances in betalain research [J]. Phytochemistry, 2003, 62(3): 247−269. doi: 10.1016/S0031-9422(02)00564-2 [3] 柳燕, 谢礼洋, 赖钟雄, 等. 苋菜amaAG基因克隆与生物信息学分析 [J]. 江西农业大学学报, 2017, 39(1):168−174. doi: 10.13836/j.jjau.2017022LIU Y, XIE L Y, LAI Z X, et al. Cloning and bioinformatics analysis of ama AG in Amaranthus tricolor L [J]. Acta Agriculturae Universitatis Jiangxiensis, 2017, 39(1): 168−174.(in Chinese) doi: 10.13836/j.jjau.2017022 [4] CUBAS P, LAUTER N, DOEBLEY J, et al. The TCP domain: A motif found in proteins regulating plant growth and development [J]. The Plant Journal, 1999, 18(2): 215−222. doi: 10.1046/j.1365-313X.1999.00444.x [5] HUO Y Z, XIONG W D, SU K L, et al. Genome-wide analysis of the TCP gene family in switchgrass (Panicum virgatum L.)[J]. International Journal of Genomics, 2019: https://doaj.org/article/613044632d6d4e2f9f696c52793c0ebd. [6] PARK H C, KIM M L, KANG Y H, et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor [J]. Plant Physiology, 2004, 135(4): 2150−2161. doi: 10.1104/pp.104.041442 [7] 李佳皓, 谢敏秋, 万传银, 等. 马铃薯转录因子StTCP13基因的原核表达及盐胁迫功能研究 [J]. 华北农学报, 2021, 36(2):33−39. doi: 10.7668/hbnxb.20191507LI J H, XIE M Q, WAN C Y, et al. Prokaryotic expression of transcription factor StTCP13 in potato and its function of salt stress [J]. Acta Agriculturae Boreali-Sinica, 2021, 36(2): 33−39.(in Chinese) doi: 10.7668/hbnxb.20191507 [8] FANG Y J, ZHENG Y Q, LU W, et al. Roles of miR319-regulated TCPs in plant development and response to abiotic stress [J]. The Crop Journal, 2021, 9(1): 17−28. doi: 10.1016/j.cj.2020.07.007 [9] 郑玲, 白雪婷, 李会云. 高粱TCP基因家族全基因组鉴定及表达分析 [J]. 河南农业科学, 2019, 48(10):30−36.ZHENG L, BAI X T, LI H Y. Genome-wide identification and expression analysis of TCP gene family in Sorghum bicolor L [J]. Journal of Henan Agricultural Sciences, 2019, 48(10): 30−36.(in Chinese) [10] 李旭娟, 林秀琴, 刘洪博, 等. 甘蔗TB1基因的克隆与生物信息学分析 [J]. 热带作物学报, 2015, 36(11):1978−1985. doi: 10.3969/j.issn.1000-2561.2015.11.010LI X J, LIN X Q, LIU H B, et al. Cloning and bioinformatics analysis of the TB1 gene in sugarcane [J]. Chinese Journal of Tropical Crops, 2015, 36(11): 1978−1985.(in Chinese) doi: 10.3969/j.issn.1000-2561.2015.11.010 [11] 任丽. 白桦BpTCP3基因的功能研究[D]. 哈尔滨: 东北林业大学, 2019.REN L. Function analysis of BpTCP3 gene in Betula platyphylla[D]. Harbin: Northeast Forestry University, 2019. (in Chinese) [12] DUAN A Q, WANG Y W, FENG K, et al. TCP family genes control leaf development and its responses to gibberellin in celery [J]. Acta Physiologiae Plantarum, 2019, 41(9): 153. doi: 10.1007/s11738-019-2945-3 [13] RIECHMANN J L, HEARD J, MARTIN G, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes [J]. Science, 2000, 290(5499): 2105−2110. doi: 10.1126/science.290.5499.2105 [14] 冯建英, 李立芹, 李佳皓, 等. 马铃薯TCP家族转录因子鉴定与表达模式分析 [J]. 基因组学与应用生物学, 2021, 40(S2):2756−2764. doi: 10.13417/j.gab.040.002756FENG J Y, LI L Q, LI J H, et al. Identification and expression pattern analysis of TCP family transcription factors in potato [J]. Genomics and Applied Biology, 2021, 40(S2): 2756−2764.(in Chinese) doi: 10.13417/j.gab.040.002756 [15] 杨婷, 黎成, 申佳瑜, 等. 茄子TCP基因家族全基因组的鉴定与分析[J/OL]. 生物工程学报: 1-21. [2022-07-06]. DOI: 10.13345/j. cjb. 220114.YANG T, LI C, SHEN JY, et al. Identification and analysis of the complete genome of the TCP gene family in Eggplant[J/OL]. Chinese Journal of Bioengineering: 1-21[2022-07-06]. DOI: 10.13345/j.cjb.220114. (in Chinese) [16] 关紫微, 曹希雅, 张先文, 等. 水稻TCP家族的全基因组鉴定及表达水平分析[J/OL]. 分子植物育种, 2020: 1-18. (2020-12-29). https://kns.cnki.net/kcms/detail/46.1068.S.20201229.0941.002.html.GUAN Z W, CAO X Y, ZHANG X W, et al. Genome-wide identification and expression analysis of TCP gene family in rice(Oryza sativa L. )[J/OL]. Molecular Plant Breeding, 2020: 1-18. (2020-12-29). https://kns.cnki.net/kcms/detail/46.1068.S.20201229.0941.002.html.(in Chinese) [17] 王景超, 张君, 齐云, 等. 玉米TCP家族基因的表达分析 [J]. 玉米科学, 2022, 30(1):63−68. doi: 10.13597/j.cnki.maize.science.20220109WANG J C, ZHANG J, QI Y, et al. Expression analysis on TCP family genes of maize [J]. Journal of Maize Sciences, 2022, 30(1): 63−68.(in Chinese) doi: 10.13597/j.cnki.maize.science.20220109 [18] 陈何, 王乐, 赵春丽, 等. 氮素和红蓝复合光配比对苋菜幼苗亚硝酸还原酶活性及其基因表达的影响 [J]. 中国农业大学学报, 2021, 26(8):61−71. doi: 10.11841/j.issn.1007-4333.2021.08.07CHEN H, WANG L, ZHAO C L, et al. Effects of nitrogen and red and blue light on NiR enzyme activity and gene expression in Amaranthus tricolor L. seedlings [J]. Journal of China Agricultural University, 2021, 26(8): 61−71.(in Chinese) doi: 10.11841/j.issn.1007-4333.2021.08.07 [19] 王乐, 陈何, 陈家兰, 等. 基于苋菜转录组的ARF基因家族鉴定及表达 [J]. 应用与环境生物学报, 2021, 27(1):167−176.WANG L, CHEN H, CHEN J L, et al. Identification and expression of ARF gene family based on Amaranthus tricolor L. transcriptome [J]. Chinese Journal of Applied and Environmental Biology, 2021, 27(1): 167−176.(in Chinese) [20] 孙菡笛, 薛江芝, 刘亚洁, 等. 玉米TCP转录因子家族的全基因组鉴定及表达模式分析 [J]. 分子植物育种, 2021, 19(8):2460−2471. doi: 10.13271/j.mpb.019.002460SUN H D, XUE J Z, LIU Y J, et al. Genome identification and expression pattern analysis of TCP transcription factor family in maize [J]. Molecular Plant Breeding, 2021, 19(8): 2460−2471.(in Chinese) doi: 10.13271/j.mpb.019.002460 [21] 丁宁, 李梓彰, 田文, 等. 小麦TCP基因家族的全基因组鉴定和对热胁迫的响应 [J]. 麦类作物学报, 2018, 38(10):1157−1165. doi: 10.7606/j.issn.1009-1041.2018.10.03DING N, LI Z Z, TIAN W, et al. Genome-wide identification of the wheat TCP gene family and its response to heat stress [J]. Journal of Triticeae Crops, 2018, 38(10): 1157−1165.(in Chinese) doi: 10.7606/j.issn.1009-1041.2018.10.03 [22] YIN Z J, LI Y, ZHU W D, et al. Identification, characterization, and expression patterns of TCP genes and microRNA319 in cotton [J]. International Journal of Molecular Sciences, 2018, 19(11): 3655. doi: 10.3390/ijms19113655 [23] 白永宏, 赵赞延, 魏雄博, 等. 谷子TCP转录因子家族成员鉴定与生物信息学分析 [J]. 南方农业学报, 2020, 51(6):1300−1307.BAI Y H, ZHAO Z Y, WEI X B, et al. Identification and bioinformatics analysis of TCP transcription factor family member in Setaria italic L. (foxtail millet) [J]. Journal of Southern Agriculture, 2020, 51(6): 1300−1307.(in Chinese) [24] 冯志娟, 徐盛春, 刘娜, 等. 植物TCP转录因子的作用机理及其应用研究进展 [J]. 植物遗传资源学报, 2018, 19(1):112−121.FENG Z J, XU S C, LIU N, et al. Molecular mechanisms and applications of TCP transcription factors in plants [J]. Journal of Plant Genetic Resources, 2018, 19(1): 112−121.(in Chinese) [25] 张延召, 罗新, 李会云, 等. 牡丹TCP家族基因的鉴定与生物信息学分析 [J]. 分子植物育种, 2022, 20(1):31−37. doi: 10.13271/j.mpb.020.000031ZHANG Y Z, LUO X, LI H Y, et al. Identification and bioinformatics analysis of TCP family genes in tree peony [J]. Molecular Plant Breeding, 2022, 20(1): 31−37.(in Chinese) doi: 10.13271/j.mpb.020.000031 [26] HERVÉ C, DABOS P, BARDET C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development [J]. Plant Physiology, 2009, 149(3): 1462−1477. doi: 10.1104/pp.108.126136 [27] 何志敏. 拟南芥转录因子TCP2与CRY1相互作用的遗传学与生化分析[D]. 长沙: 湖南大学, 2016.HE Z M. The molecular genetical and biochemical study of transcription factor TCP2 and CRY1 interaction in Arabidopsis[D]. Changsha: Hunan University, 2016. (in Chinese) [28] 安新艳, 楼盼盼, 郝娟. 植物TCP转录因子的研究进展 [J]. 安徽农业科学, 2020, 48(15):20−23,27. doi: 10.3969/j.issn.0517-6611.2020.15.006AN X Y, LOU P P, HAO J. Research progress on plant TCP transcription factors [J]. Journal of Anhui Agricultural Sciences, 2020, 48(15): 20−23,27.(in Chinese) doi: 10.3969/j.issn.0517-6611.2020.15.006 [29] 安家兴. 拟南芥TCP11调控维管束的发育[D]. 兰州: 兰州大学, 2012.AN J X. TCP11Regalates the development of vascular bundles in arahidopsis thaliana[D]. Lanzhou: Lanzhou University, 2012. (in Chinese) [30] KOROLEVA O A, TOMLINSON M L, LEADER D, et al. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions [J]. The Plant Journal:for Cell and Molecular Biology, 2005, 41(1): 162−174. [31] 张松莲, 曾富华, 喻宁华, 等. 植物miRNA的功能及其作用机制 [J]. 热带亚热带植物学报, 2006, 14(5):444−450. doi: 10.3969/j.issn.1005-3395.2006.05.015ZHANG S L, ZENG F H, YU N H, et al. Advances in studies on the function and mechanism of plant microRNA [J]. Journal of Tropical and Subtropical Botany, 2006, 14(5): 444−450.(in Chinese) doi: 10.3969/j.issn.1005-3395.2006.05.015 [32] 李坤杰, 谭杉杉, 孙勃, 等. 芥菜TCP转录因子家族全基因组鉴定及表达分析 [J]. 四川农业大学学报, 2019, 37(4):459−468.LI K J, TAN S S, SUN B, et al. Genome-wide identification and analysis of TCP transcription factor family in Brassica juncea [J]. Journal of Sichuan Agricultural University, 2019, 37(4): 459−468.(in Chinese) [33] LIN Y F, CHEN Y Y, HSIAO Y Y, et al. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris [J]. Journal of Experimental Botany, 2016, 67(17): 5051−5066. doi: 10.1093/jxb/erw273 [34] KOYAMA T, SATO F, OHME-TAKAGI M. Roles of miR319 and TCP transcription factors in leaf development [J]. Plant Physiology, 2017, 175(2): 874−885. doi: 10.1104/pp.17.00732 [35] LIU S C, ZHENG X L, PAN J F, et al. RNA-sequencing analysis reveals betalains metabolism in the leaf of Amaranthus tricolor L [J]. PLoS One, 2019, 14(4): e0216001. doi: 10.1371/journal.pone.0216001 [36] 王廷芹, 甘秋霞, 李倩茹. 盐胁迫对苋菜种子的发芽及幼苗生长的影响 [J]. 贵州大学学报(自然科学版), 2021, 38(1):10−15,32. doi: 10.15958/j.cnki.gdxbzrb.2021.01.02WANG T Q, GAN Q X, LI Q R. Effect of salt stress on seed germination and seedling growth of amaranth [J]. Journal of Guizhou University (Natural Sciences), 2021, 38(1): 10−15,32.(in Chinese) doi: 10.15958/j.cnki.gdxbzrb.2021.01.02 [37] 曾泳怡, 冯梓晴, 曾晓靖, 等. NaCl胁迫对不同基因型水稻种子萌发和幼苗生长的影响 [J]. 安徽农业科学, 2021, 49(7):25−29. doi: 10.3969/j.issn.0517-6611.2021.07.007ZENG Y Y, FENG Z Q, ZENG X J, et al. Effects of NaCl stress on seed germination and seedling growth of different rice genotypes [J]. Journal of Anhui Agricultural Sciences, 2021, 49(7): 25−29.(in Chinese) doi: 10.3969/j.issn.0517-6611.2021.07.007 -







 下载:
下载: