Transcriptome Sequencing and Identification of Genes Associated with Flavonoid Biosynthesis in Stellaria yunnanensis Roots
-
摘要:
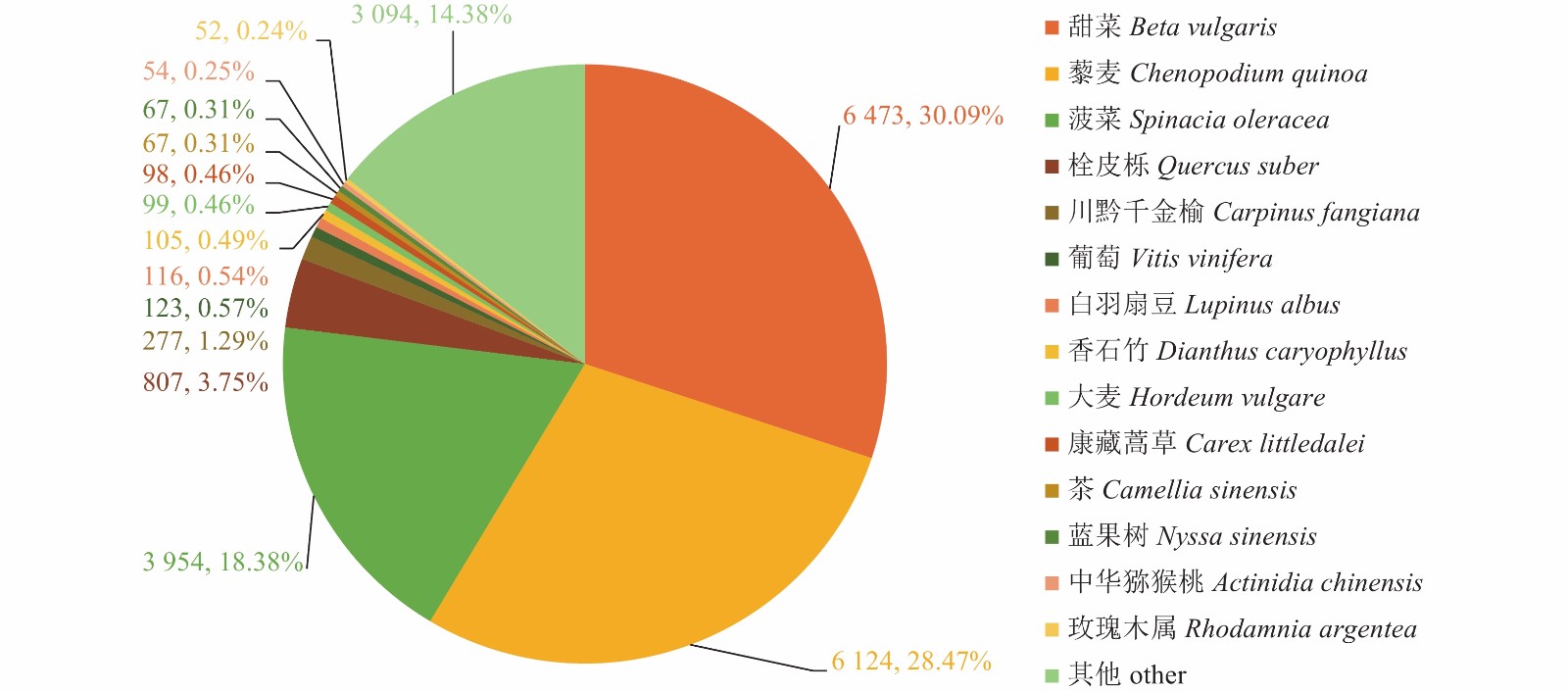
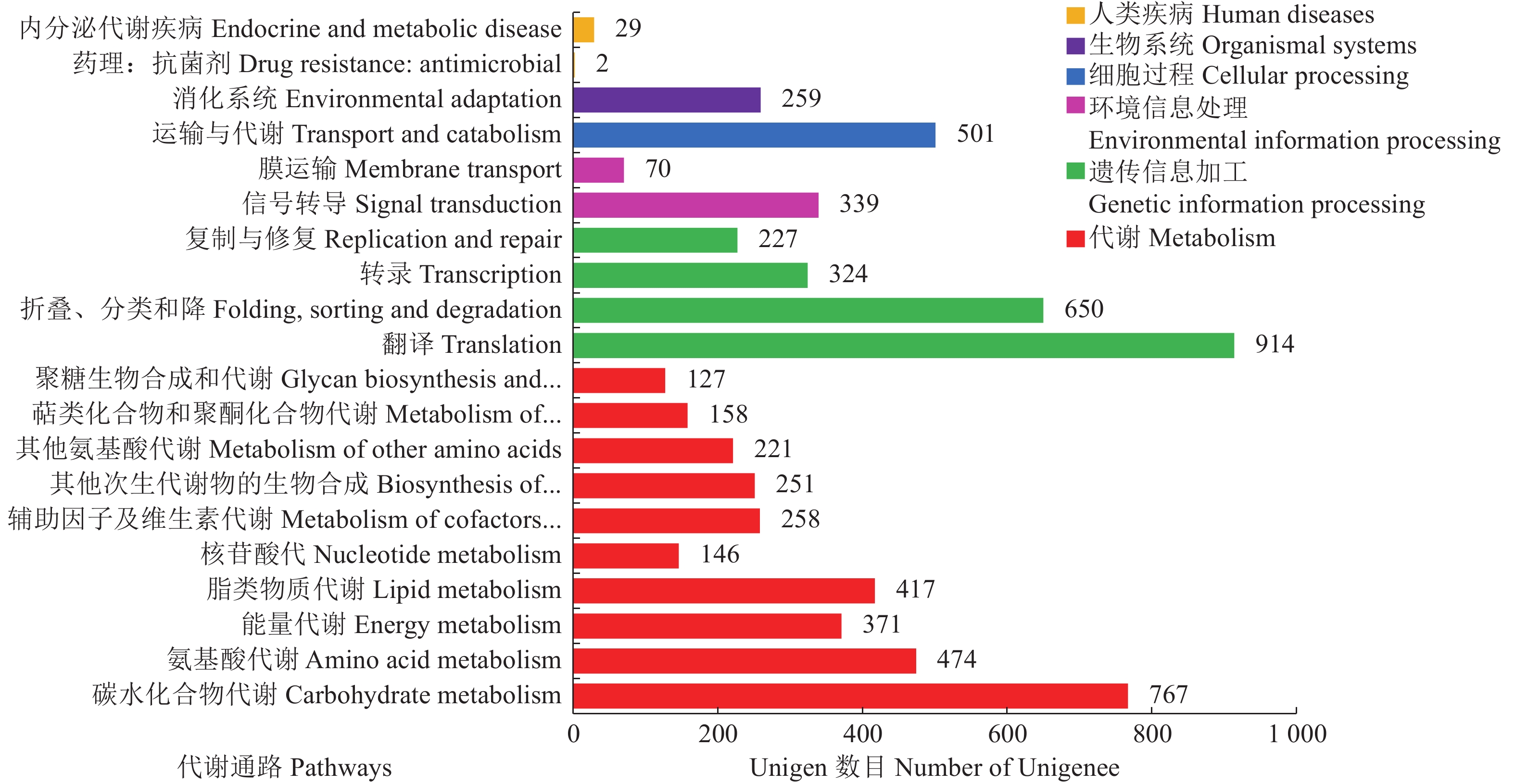
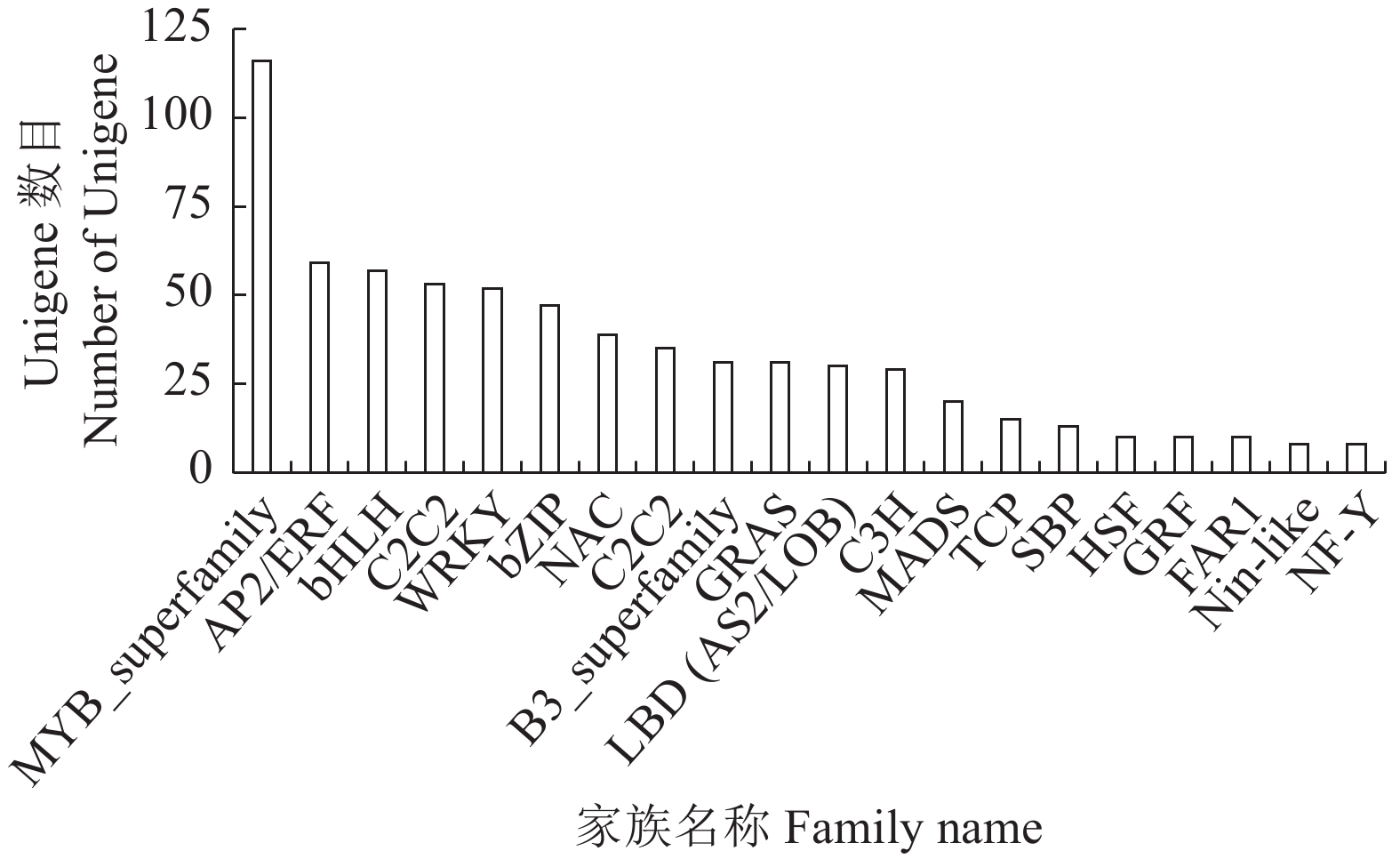
目的 通过高通量测序获取千针万线草根转录组信息,在分子水平上进一步研究千针万线草根黄酮类的生物合成。 方法 采用高通量测序技术平台lllumina Novaseq 6000完成千针万线草根的转录组测序,对unigenes进行功能注释和解析黄酮类化合物的生物合成相关基因。 结果 总共得到34137条Unigenes,平均长度为1093.58 bp。注释到六大功能数据库(NR、egg NOG、Pfam、Swiss-Prot、GO、KEGG)中的Unigenes总数达到22369条。千针万线草根Unigenes匹配至NR数据库的有21510条,与黎科的甜菜、藜麦和菠菜有高度同源性;19414条Unigenes在egg NOG数据库得到19980个注释并划分为23类;19942条Unigenes在GO数据库中获得69356个注释,根据功能划分为细胞组分、分子功能及生物过程三大类,分别对应14、16、23个亚类,其中涉及生物过程较多;6505条Unigenes富集在KEGG数据库的131条通路中,代谢相关的通路占比最大,其中筛选获得80个与黄酮类化合物代谢相关的基因,共编码16个关键酶。同时,有724个Unigenes被注释为转录因子。 结论 对千针万线草根进行转录组测序及基因功能注释,并筛选到多个与黄酮类化合物合成相关的Unigenes,研究结果丰富了千针万线草根的遗传信息,可以为进一步鉴定该物种药用成分合成的关键基因及其调控机制提供一定的参考依据。 Abstract:Objective Transcriptome of Stellaria yunnanensis root was sequenced and genes associated with the flavonoid biosynthesis identified. Method Transcriptome of S. yunnanensis root was sequenced using the high-throughput Illumina Novaseq 6 000. Functional annotation of the unigenes were conducted and genes related to the biosynthesis identified. Result There were 34 137 unigenes found with an average length of 1 093.58 bp. They annotated to 6 functional databases, including NR, egg NOG, Pfam, Swiss-Prot, GO, and KEGG, that numbered 22 369 in total. There were 21 510 unigenes annotated in NR database, which had high homology with Beta vulgaris, Chenopodium quinoa, and Spinacia oleracea of Chenopodiaceae, while 19 980 from 19 414 unigenes in egg NOG in 23 categories, and 69 356 from 19 942 unigenes in GO in 3 major functional categories of cellular components, molecular functions, and, especially, biological processes with 14, 16, and 23 subcategories, respectively. Of the unigenes, 6 505 were enriched in 131 pathways in KEGG with the largest proportion related to the metabolism pathways, 80 associated with the flavonoid biosynthesis encoding 16 key enzymes, and 724 annotated as transcription factors. Conclusion The high-throughput transcriptome sequencing and gene function annotation on the S. yunnanensis roots were successfully performed. Several unigenes related to the flavonoid synthesis were identified. The information would facilitate further studies on the key genes and mechanisms in the formation of medicinal important functional ingredient in the plant. -
Key words:
- Stellaria yunnanensis /
- root /
- transcriptome /
- sequencing /
- gene functional annotation
-
图 3 千针万线草unigene的egg NOG分类
A−RNA加工与修饰;B−染色质结构与动力;C−能量生产与转化;D−细胞周期调控、细胞分裂、染色体分裂;E−氨基酸转运与代谢;F−核苷酸转运与代谢;G−糖转运与代谢;H−辅酶转运与代谢;I−脂类转运与代谢;J−翻译、核糖体结构和生物合成;K−转录;L−复制、重组和修复;M−细胞壁、膜、包体生物合成;N−细胞运动性;O−蛋白质翻译后修饰与周转;P−无机离子转运与代谢;Q−次生代谢产物的生物合成; S−未知功能;T−信号传导机制;U−胞内运输、分泌和囊泡运输;V−防御机制; Y−核酸结构;Z−细胞骨架。
Figure 3. Egg NOG function classification of S. yunnanensis
A: RNA processing and modification; B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination and repair; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; O: Posttranslational modification, protein turnover, chaperones; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms; Y: Nuclear structure; Z: Cytoskeleton.
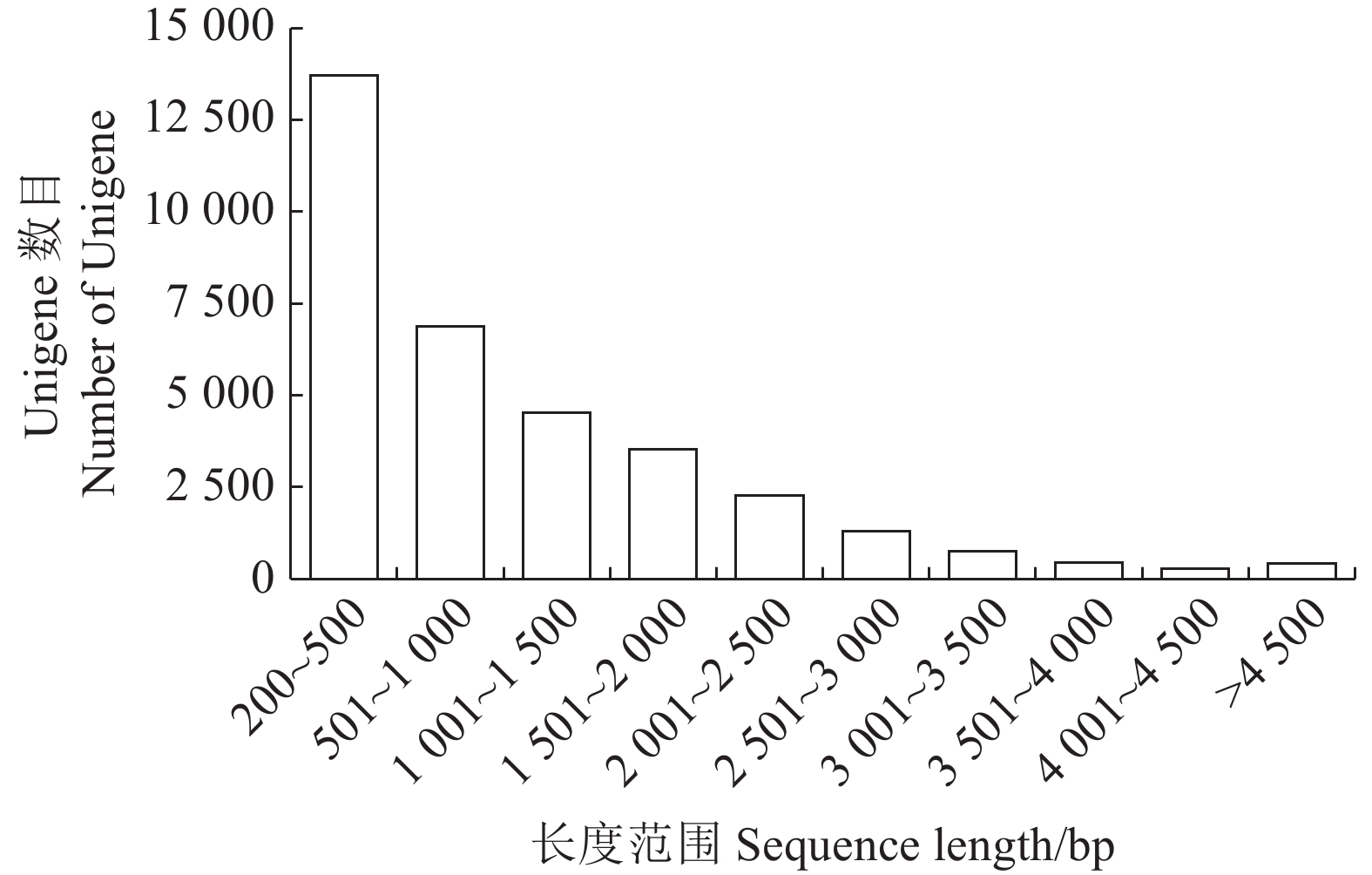
表 1 Unigene基本信息表
Table 1. Basic information on unigenes
基因数量
Genes NumGC核苷酸含量
GC Percentage/%N50/bp 最大长度
Max Length/bp最小长度
Min Length/bp平均长度
Average Length/bp组装的总碱基
Total assembled bases34137 41.02 1799 14683 201 1093.58 37331463 表 2 Unigene注释统计
Table 2. Annotation of unigenes
注释数据库
Annotation databaseUnigene数目
Number of Unigene比例
Percentage /%Nr 21510 63.01 Swiss-prot 17208 50.41 Pfam 17702 51.86 egg NOG 19414 56.87 GO 17191 50.36 KEGG 10285 30.13 注释到的基因数量Annotation genes 22369 65.53 未注释到的基因数量Without annotation gene 11768 34.47 表 3 千针万线草根转录组中黄酮类化合物合成相关基因
Table 3. Flavonoid biosynthesis-related gene in transcriptome of S. yunnanensis roots
代谢途径
Metabolic pathway基因名称
GeneKO编号
KO IDEC编号
EC No.Unigene数量
Number of unigene苯丙烷代谢通路
Phenylpropane metabolic pathway苯丙氨酸解氨酶基因(PAL) K10775 4.3.1.24 5 肉桂酸-4-单加氧酶基因(CYP73A) K00487 1.14.14.91 5 CoA连接酶基因(4CL) K01904 6.2.1.12 8 类黄酮代谢通路
Flavonoid metabolic pathway花青素还原酶基因(ANR) K08695 1.3.1.77 1 香豆酰脂3’羟化酶基因(C3’H) K09754 1.11.13.36 2 莽草酸羟基肉桂转移酶基因(HCT) K13065 2.3.1.133 12 查耳酮异构酶基因(CHI) K01859 5.5.1.6 3 肉桂酸-4-单加氧酶基因(CYP73A) K00487 1.14.14.91 5 根皮苷合酶基因(PGT1) K22845 2.4.1.357 1 查尔酮合成酶基因(CHS) K00660 2.3.1.74 12 咖啡酰辅酶A-O甲基转移酶基因(CCOAOMT) K00588 2.1.1.104 3 类黄酮-3’单加氧酶基因(CYP75B1) K05280 1.14.14.82 4 黄烷酮-3-羟化酶基因(F3H) K00475 1.14.11.9 2 黄酮醇代谢通路
Flavonol metabolic pathway类黄酮-3’单加氧酶基因(CYP75B1) K05280 1.14.14.82 4 黄酮醇-3-0-葡萄糖苷葡萄糖基转移酶基因(FG3) K22794 2.4.1.239 6 异黄酮代谢通路
Isoflavone metabolic pathway异黄酮2’-羟化酶基因(CYP81E) K13260 1.14.14.90 7 -
[1] 中国植物志编委会. 中国植物志(第26卷) [M]. 北京: 科学出版社, 1996: 133. [2] 马利华, 陈斌, 谢东浩, 等. 繁缕属植物抗病毒活性物质基础组分结构研究 [J]. 中草药, 2012, 43(4):799−805.MA L H, CHEN B, XIE D H, et al. Study on antiviral material basis and component structure in plants of Stellaria L [J]. Chinese Traditional and Herbal Drugs, 2012, 43(4): 799−805.(in Chinese) [3] 谭瑞璞, 刘海周, 邓义德, 等. 千针万线草中总黄酮提取工艺的研究 [J]. 云南中医学院学报, 2009, 32(1):9−11. doi: 10.3969/j.issn.1000-2723.2009.01.003TAN R P, LIU H Z, DENG Y D, et al. Study on extraction process of total flavonoids in Stellaria yunnanensis franch [J]. Journal of Yunnan University of Traditional Chinese Medicine, 2009, 32(1): 9−11.(in Chinese) doi: 10.3969/j.issn.1000-2723.2009.01.003 [4] 谭瑞璞, 刘海周, 梁晓原. HPLC法测定千针万线草中牡荆素的含量 [J]. 云南中医学院学报, 2009, 32(4):28−29,32. doi: 10.3969/j.issn.1000-2723.2009.04.009TAN R P, LIU H Z, LIANG X Y. Determination of vitexin in Stellaria yunnanensis by HPLC [J]. Journal of Yunnan University of Traditional Chinese Medicine, 2009, 32(4): 28−29,32.(in Chinese) doi: 10.3969/j.issn.1000-2723.2009.04.009 [5] 刘伟, 王俊燚, 李萌, 等. 基于转录组测序的银杏类黄酮生物合成关键基因表达分析 [J]. 中草药, 2018, 49(23):5633−5639. doi: 10.7501/j.issn.0253-2670.2018.23.024LIU W, WANG J Y, LI M, et al. Transcriptome sequencing analysis of gene expression of flavonoid biosynthesis in Ginkgo biloba [J]. Chinese Traditional and Herbal Drugs, 2018, 49(23): 5633−5639.(in Chinese) doi: 10.7501/j.issn.0253-2670.2018.23.024 [6] 邹丽秋, 王彩霞, 匡雪君, 等. 黄酮类化合物合成途径及合成生物学研究进展 [J]. 中国中药杂志, 2016, 41(22):4124−4128.ZOU L Q, WANG C X, KUANG X J, et al. Advance in flavonoids biosynthetic pathway and synthetic biology [J]. China Journal of Chinese Materia Medica, 2016, 41(22): 4124−4128.(in Chinese) [7] LI H Y, DONG Y Y, YANG J, et al. De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids [J]. PLoS One, 2012, 7(2): e30987. doi: 10.1371/journal.pone.0030987 [8] 邓楠, 史胜青, 常二梅, 等. 基于中麻黄萌发种子转录组的黄酮类化合物合成途径基因的挖掘 [J]. 林业科学研究, 2014, 27(6):758−763.DENG N, SHI S Q, CHANG E M, et al. Exploring flavonoid biosynthetic pathway genes based on transcriptome of Ephedra intermedia germinating seeds [J]. Forest Research, 2014, 27(6): 758−763.(in Chinese) [9] 邹毅辉, 刘昌勇, 林泽燕, 等. 半枝莲转录组高通量测序及黄酮类相关基因解析 [J]. 福建农业学报, 2018, 33(12):1242−1250.ZOU Y H, LIU C Y, LIN Z Y, et al. Transcriptome sequencing and flavonoid biosynthesis-related genes of Scutellaria barbata D. Don [J]. Fujian Journal of Agricultural Sciences, 2018, 33(12): 1242−1250.(in Chinese) [10] 邹福贤, 许文, 黄泽豪, 等. 金线莲转录组测序及其黄酮类合成相关基因分析 [J]. 中国药科大学学报, 2019, 50(1):66−74. doi: 10.11665/j.issn.1000-5048.20190109ZOU F X, XU W, HUANG Z H, et al. Analysis of transcriptome sequencing and related genes of flavonoid biosynthesis from Anoectochilus roxburghii [J]. Journal of China Pharmaceutical University, 2019, 50(1): 66−74.(in Chinese) doi: 10.11665/j.issn.1000-5048.20190109 [11] 林江波, 王伟英, 邹晖, 等. 基于转录组测序的铁皮石斛黄酮代谢途径及相关基因解析 [J]. 福建农业学报, 2019, 34(9):1019−1025.LIN J B, WANG W Y, ZOU H, et al. Transcriptome analysis on pathway of and genes related to flavonoid synthesis in Dendrobium officinale [J]. Fujian Journal of Agricultural Sciences, 2019, 34(9): 1019−1025.(in Chinese) [12] 许明, 杨志坚, 黄学敏, 等. 藤茶高通量转录组分析及黄酮类化合物合成相关基因挖掘 [J]. 南方农业学报, 2020, 51(8):1797−1805.XU M, YANG Z J, HUANG X M, et al. Transcriptome analysis of Ampelopsis grossedentata(Hand. Mazz. )W. T. Wang and mining of putative genes involved in flavonoid biosynthesis [J]. Journal of Southern Agriculture, 2020, 51(8): 1797−1805.(in Chinese) [13] 袁茂, 江明锋, 徐亚欧, 等. 藏鸡不同发育阶段腿部肌肉组织转录组及microRNA联合分析 [J]. 畜牧兽医学报, 2019, 50(12):2400−2412. doi: 10.11843/j.issn.0366-6964.2019.12.004YUAN M, JIANG M F, XU Y O, et al. Analysis of transcriptome and microRNA in leg muscle of Tibetan chicken at different developmental stages [J]. Acta Veterinaria et Zootechnica Sinica, 2019, 50(12): 2400−2412.(in Chinese) doi: 10.11843/j.issn.0366-6964.2019.12.004 [14] GRABHERR M G, HAAS B J, YASSOUR M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome [J]. Nature Biotechnology, 2011, 29(7): 644−652. doi: 10.1038/nbt.1883 [15] SMITH-UNNA R, BOURSNELL C, PATRO R, et al. TransRate: Reference-free quality assessment of de novo transcriptome assemblies [J]. Genome Research, 2016, 26(8): 1134−1144. doi: 10.1101/gr.196469.115 [16] LI W Z, GODZIK A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences [J]. Bioinformatics, 2006, 22(13): 1658−1659. doi: 10.1093/bioinformatics/btl158 [17] SIMÃO F A, WATERHOUSE R M, IOANNIDIS P, et al. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs [J]. Bioinformatics, 2015, 31(19): 3210−3212. doi: 10.1093/bioinformatics/btv351 [18] CONESA A, GÖTZ S, GARCÍA-GÓMEZ J M, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research [J]. Bioinformatics, 2005, 21(18): 3674−3676. doi: 10.1093/bioinformatics/bti610 [19] 朱畇昊, 董诚明, 郑晓珂, 等. 基于转录组测序的山茱萸次生代谢生物合成相关基因的挖掘 [J]. 中国中药杂志, 2017, 42(2):213−219.ZHU Y H, DONG C M, ZHENG X K, et al. Transcriptome analysis reveals genes involved in biosynthesis of secondary metabolism in Cornus officinalis [J]. China Journal of Chinese Materia Medica, 2017, 42(2): 213−219.(in Chinese) [20] 吴萍, 郭俊霞, 王晓宇, 等. 基于高通量测序技术的杭白芷(Angelica dahurica)根转录组数据分析 [J]. 分子植物育种, 2020, 18(10):3207−3216.WU P, GUO J X, WANG X Y, et al. High-throughput transcriptome sequencing of roots of Angelica dahurica and data analyses [J]. Molecular Plant Breeding, 2020, 18(10): 3207−3216.(in Chinese) [21] 王建秋, 王晓丽, 曹子林. 滇白前种子转录组测序及功能注释 [J]. 种子, 2021, 40(3):52−59.WANG J Q, WANG X L, CAO Z L. Transcriptome sequencing and functional annotation of Silene viscidula seeds [J]. Seed, 2021, 40(3): 52−59.(in Chinese) [22] WANG H Y, WANG H L, SHAO H B, et al. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology [J]. Frontiers in Plant Science, 2016, 7: 67. -







 下载:
下载: