Diversity and Isolation of Symbiotic Bacteria on Delia antiqua Larvae
-
摘要:
目的 探索蒜蛆幼虫伴生细菌的多样性,并比较两种常用培养基对其伴生细菌的分离效果,为开展蒜蛆伴生细菌研究提供技术指导。 方法 采用高通量扩增子测序方法检测蒜蛆幼虫体表和肠道的细菌多样性;使用LB和TSB从蒜蛆幼虫的体表和肠道分离细菌,并比较两种培养基分离结果、测序结果和两种培养基分离效果的差异。 结果 高通量测序从蒜蛆幼虫体表和肠道共检测到4个门、66个科、115个属的细菌,其中绝大部分不能够鉴定到种。使用LB培养基从蒜蛆幼虫体表和肠道共分离得到463株细菌,鉴定后共得到29种细菌,分别属于11个科18个属。使用TSB培养基从蒜蛆幼虫体表和肠道分离得到391株细菌,经测序后共鉴定到44种细菌,分别属于19个科28个属。总体来说,TSB分离到的细菌种类更多,部分细菌可同时用LB与TSB分离到;此外,与高通量测序结果相比,两种培养基均只能分离到很小一部分种类的细菌。 结论 在蒜蛆伴生细菌的分离试验中,最好同时使用两种培养基对细菌进行分离,以获得更多种类的细菌。 Abstract:Objective Diversity of the symbiotic bacteria on Delia antiqua larvae was studied, and effectiveness of two commonly used culture media in isolating them compared. Methods Diversity of the bacterial population on the surface and in the guts of D. antiqua larvae were determined using the high-throughput amplicon sequencing technique. The effectiveness of the LB and TSB media to culture and isolate the bacteria were compared based on the sequencing results. Results The inhabitation bacteria on the larvae were from 115 genera, 66 families, and 4 phyla according to the sequencing. Most of them could not be annotated to species level. On LB medium, 463 strains were isolated that belonged to 29 species, 18 genera, and 11 families, while on TSB, 391 strains from 44 species, 28 genera, and 19 families. TSB isolated more species than LB, and some of the species could be isolated by either method. However, the high-throughput amplicon sequencing revealed that these culture methods could provide only a minute portion of the bacteria population that coinhabited on the host. Conclusion It was necessary to use both TSB and LB media in order to maximize the isolation of all symbiotic bacteria on D. antiqua larvae. -
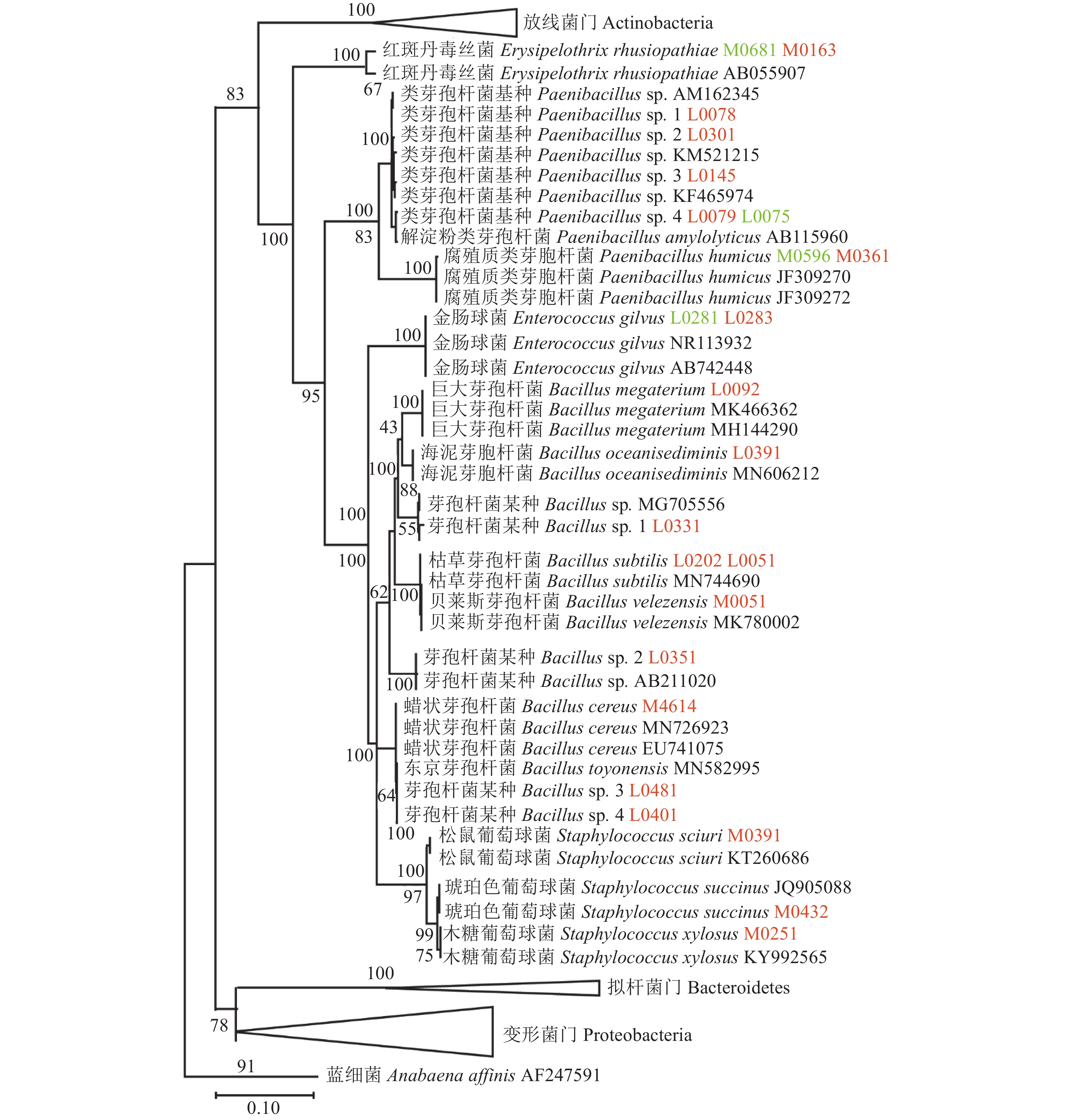
图 3 两种培养基分离蒜蛆幼虫细菌的16S rDNA最大似然树
注:图中L、M开头的菌株为LB、TSB培养基分离得到的;其中标为橙色、绿色的菌株为从蒜蛆幼虫体表和肠道分离得到的,下同。
Figure 3. Maximum likelihood trees for 16S rDNA sequences of strains on D. antiqua larva isolated on two media
Note: Strains numbered L and M are isolations from LB and TSB media, respectively. Orange and green indicate strains from surface and guts of D. antiqua larvae, respectively. Same for below.
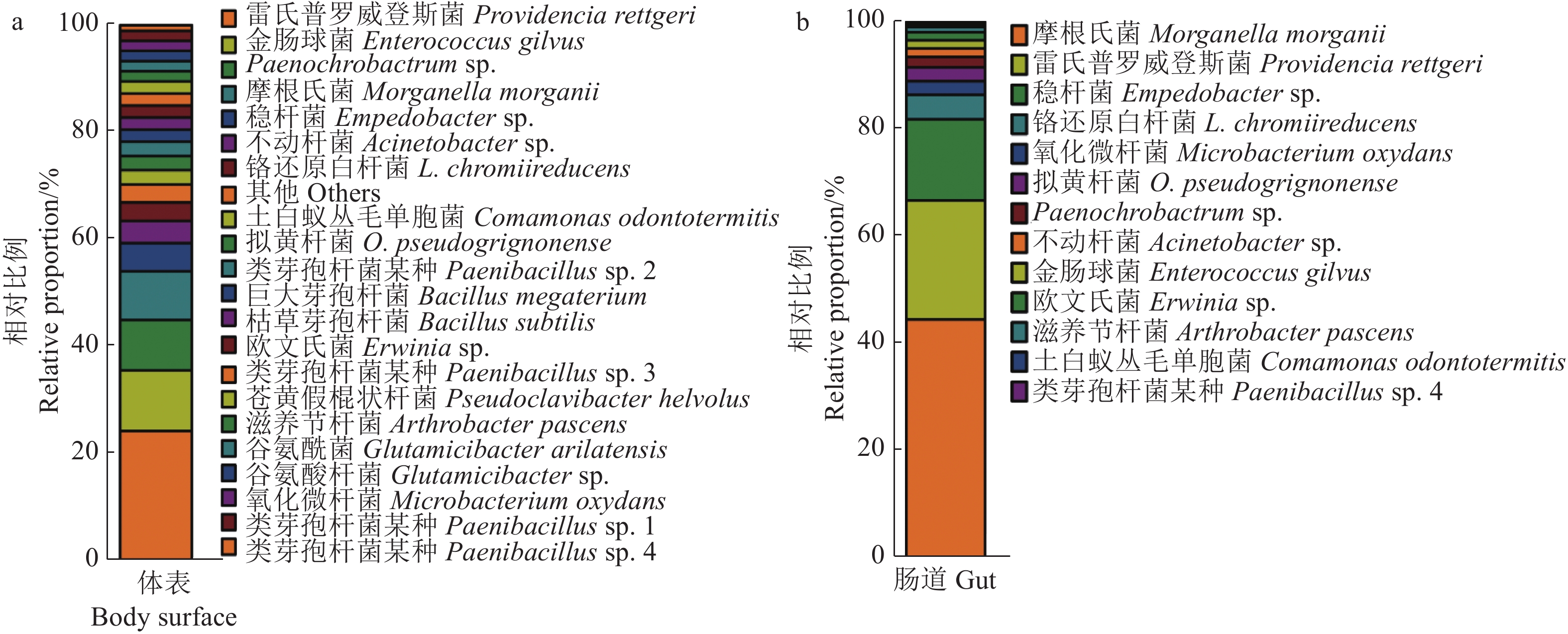
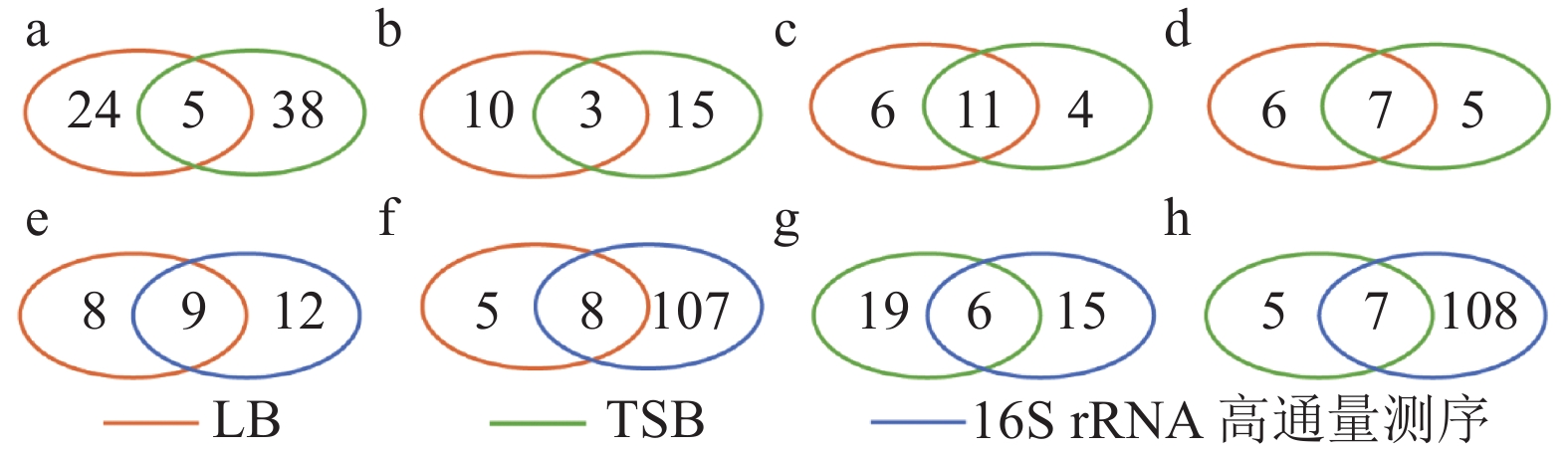
图 7 蒜蛆幼虫使用LB、TSB培养基以及测序分离细菌种类比较
注:图中a、b为种水平细菌种类比较;c-h为属水平细菌种类比较;a、c、e、g为蒜蛆幼虫体表细菌种类比较;b、d、f、h为蒜蛆幼虫肠道细菌种类比较。
Figure 7. Bacteria from D. antiqua larva as identified by LB, TSB, and high throughput sequencing methods
Note: Figs. a and b represent bacteria on species level; Figs. c-h, on genus level; Figs. a, c, e, and g, from D. antiqua larval surface; and Figs. b, d, f, and h, from D. antiqua larval guts.
表 1 使用两种培养基分离得到的蒜蛆幼虫伴生细菌菌株的GenBank号以及种属鉴定信息
Table 1. GenBank codes and species identification information on bacteria associated with D. antiqua larvae isolated on two media
菌株号
Strain No.Genbank登录号
GenBank accession No.最相似菌株
Closest type strains相似率
Similarity/%M6595 OK642282 谷氨酸棒状杆菌 Corynebacterium glutamicum DQ173747.1 99.79 M4615 OL304276 红杆菌 Rhodobacter sp. MT036106.1 98.53 M4614 OK642283 蜡样芽孢杆菌 Bacillus cereus KY495215.1 99.86 M4613 OL304277 红杆菌 Rhodobacter sp. JF710650.1 96.67 M4110 OK642284 假苍白杆菌 Pseudochrobactrum sp. KF735806.1 99.92 M4061 OK642285 微小芽胞杆菌 Brevundimonas diminuta LC420058.1 99.78 M4025 OK642286 Paenochrobactrum glaciei NR112750.1 99.00 M2014 OK642287 拟黄杆菌 O. pseudogrignonensis MK517588.1 99.93 M0685 OK642288 食酸菌 Acidovorax sp. AB646315.1 99.79 M0684 OK642289 肺炎克雷氏伯杆菌 Acinetobacter guillouiae KF873017.1 98.63 M0681 OK642290 红斑丹毒丝菌 Erysipelothrix rhusiopathiae MN933918.1 97.61 M0678 OK642291 摩根氏菌 Morganella morganii HQ407316.1 99.18 M0662 OK642292 亮杆菌某种 Leucobacter sp. GQ344412.1 99.42 M0596 OK642293 腐殖质类芽胞杆菌 Paenibacillus humicus AM411529.2 99.73 M0564 OK642294 不动杆菌某种 Acinetobacter sp. KT261003.1 99.71 M0545 OK642295 溶血假单胞菌 P. asaccharolyticum KU597492.1 99.79 M0512 OK642296 产酸克雷伯菌 Klebsiella oxytoca LC049195.1 99.65 M0497 OK642297 微杆菌某种 Microbacterium sp. KR906366.1 99.78 M0483 OK642298 节杆菌某种 Arthrobacter sp. JX566636.1 99.72 M0432 OK642299 琥珀色葡萄球菌 Staphylococcus succinus GU084442.1 100.00 M0419 OK642300 人苍白杆菌 Ochrobactrum anthropi MG897139.1 99.85 M0417 OK642301 微杆菌某种 Microbacterium sp. LT009507.1 99.93 M0351 OK642302 俊片菌 Lampropedia sp. MK733277.1 99.28 M0332 OK642303 氧化硫副球菌 Paracoccus sulfuroxidans JQ291588.1 99.63 M0321 OK642304 雷氏普罗威登斯菌 Providencia rettgeri MN006177.1 99.73 M0291 OK642305 苍黄假棍状杆菌 Pseudoclavibacter helvolus MG694511.1 99.86 M0281 OK642306 马红球菌 Rhodococcus equi LC020105.1 99.86 M0251 OK642307 木糖葡萄球菌 Staphylococcus xylosus KT260686.1 99.59 M0211 OK642308 节杆菌某种 Arthrobacter sp. MK087736.1 99.78 M0207 OK642309 干丘亮杆菌 Leucobacter aridicollis NR 042288.1 99.65 M0201 OK642310 铬还原白杆菌 Leucobacter chromiireducens NR042287.1 99.58 M0181 OK642311 鞘氨醇杆菌 Sphingobacterium sp. NR159136.1 100.00 M0144 OK642312 酯香微杆菌 M. esteraromaticum NR026468.1 99.44 M0126 OK642313 亮杆菌某种 Leucobacter sp. KP340979.1 100.00 M0064 OK642314 粪产碱菌 Alcaligenes faecalis MK968769.1 99.86 M0055 OK642315 血杆菌某种 Sanguibacter sp. JX997905.1 99.79 M0054 OK642316 微杆菌某种 Microbacterium sp. EU841532.1 99.09 M0051 OK642317 贝莱斯芽孢杆菌 Bacillus velezensis MK780002.1 100.00 M0049 OK642318 短稳杆菌 Empedobacter brevis MN596020.1 99.93 M0047 OK642319 气味类香菌 Myroides odoratus JQ407801.1 99.85 M0021 OK642320 亮杆菌某种 Leucobacter sp. MN099406.1 97.56 L5111 OK642321 雷氏普罗威登斯菌 Providencia rettgeri MN006177.1 99.66 L0506 OK642322 摩根氏菌 Morganella morganii HQ407316.1 99.81 L0505 OK642323 土白蚁丛毛单胞菌 Comamonas odontotermitis NR 043859.1 99.72 L0502 OK605023 氧化微杆菌 Microbacterium oxydans MF459692.1 99.79 L0501 OK642324 铬还原白杆菌 Leucobacter chromiireducens NR 042287.1 99.93 L0491 OK642325 微杆菌某种 Microbacterium sp. LC133735.2 99.86 L0481 OK642326 芽孢杆菌某种 Bacillus sp. MN590524.1 99.80 L0401 OK642327 芽孢杆菌某种 Bacillus sp. MN582995.1 99.80 L0391 OK642328 海泥芽胞杆菌 Bacillus oceanisediminis MN606212.1 100.00 L0361 OK642329 稳杆菌 Empedobacter sp. MK224650.1 99.79 L0351 OK642330 芽孢杆菌某种 Bacillus sp. AB211009.1 99.80 L0331 OK642331 芽孢杆菌某种 Bacillus sp. AM913937.1 99.39 L0301 OK642332 类芽胞杆菌某种 Paenibacillus sp. KM521215.1 99.53 L0294 OK642333 铬还原白杆菌 Leucobacter chromiireducens NR 042287.1 99.93 L0281 OK642334 金肠球菌 Enterococcus gilvus NR 043793.1 99.93 L0271 OK642335 稳杆菌 Empedobacter sp. MK224650.1 99.79 L0267 OK642336 雷氏普罗威登斯菌 Providencia rettgeri MN006177.1 99.66 L0221 OK642337 谷氨酰菌 Glutamicibacter arilaitensis MG788347.1 99.93 L0202 OK642338 枯草芽孢杆菌 Bacillus subtilis KX901787.1 100.00 L0192 OK642339 谷氨酰菌 Glutamicibacter arilaitensis MG788347.1 99.93 L0182 OK642340 土白蚁丛毛单胞菌 Comamonas odontotermitis NR 043859.1 99.72 L0166 OK642341 摩根氏菌 Morganella morganii HQ407316.1 99.18 L0145 OK642342 类芽胞杆菌某种 Paenibacillus sp. KR055030.1 99.59 L0138 OK642343 拟黄杆菌 O. pseudogrignonensis KC342237.1 98.64 L0119 OK642344 欧文氏菌 Erwinia sp. KP836251.1 99.38 L0092 OK642345 巨大芽孢杆菌 Bacillus megaterium MH144290.1 99.86 L0081 OK642346 Paenochrobactrum sp. MG596953.1 98.81 L0079 OK642347 类芽胞杆菌某种 Paenibacillus sp. KF465974.1 99.59 L0078 OK642348 类芽胞杆菌某种 Paenibacillus sp. AM162345.1 99.52 L0062 OK642349 谷氨酸杆菌 Glutamicibacter sp. MH725567.1 99.93 L0051 OK642350 枯草芽孢杆菌 Bacillus subtilis KX901787.1 100.00 L0042 OK642351 滋养节杆菌 Arthrobacter pascens GQ360068.1 98.96 L0031 OK642352 欧文氏菌 Erwinia sp. KP836251.1 99.38 L0021 OK642353 皮特不动杆菌 Acinetobacter pittii MN490071.1 99.86 L0013 OK642354 溶血假单胞菌 P. asaccharolyticum MF285796.1 98.08 L0011 OK605542 苍黄假棍状杆菌 Pseudoclavibacter helvolus NR 029264.1 99.93 -
[1] NAAZ N, CHOUDHARY J S, PRABHAKAR C S, et al. Identification and evaluation of cultivable gut bacteria associated with peach fruit fly, Bactrocera zonata (Diptera: Tephritidae) [J]. Phytoparasitica, 2016, 44(2): 165−176. doi: 10.1007/s12600-016-0518-1 [2] LIU L J, MARTINEZ S I, MAZZON L, et al. Bacterial communities associated with invasive populations of Bactrocera dorsalis (Diptera: Tephritidae) in China [J]. Bulletin of Entomological Research, 2016, 106(6): 718−728. doi: 10.1017/S0007485316000390 [3] LI K L, CHEN H Y, JIANG J J, et al. Diversity of bacteriome associated with Phlebotomus chinensis (Diptera: Psychodidae) sand flies in two wild populations from China [J]. Scientific Reports, 2016, 6: 36406. doi: 10.1038/srep36406 [4] DOUGLAS A E. Multiorganismal insects: Diversity and function of resident microorganisms [J]. Annual Review of Entomology, 2015, 60: 17−34. doi: 10.1146/annurev-ento-010814-020822 [5] VERAS F F, CORREA A P F, WELKE J E, et al. Inhibition of mycotoxin-producing fungi by Bacillus strains isolated from fish intestines [J]. International Journal of Food Microbiology, 2016, 238: 23−32. doi: 10.1016/j.ijfoodmicro.2016.08.035 [6] BANSAL R, HULBERT S, SCHEMERHORN B, et al. Hessian fly-associated bacteria: transmission, essentiality, and composition [J]. Plos One, 2011, 6(8): e23170. doi: 10.1371/journal.pone.0023170 [7] MCCUTCHEON J P, MORAN N A. Extreme genome reduction in symbiotic bacteria [J]. Nature Reviews Microbiology, 2011, 10(1): 13−26. [8] MORALES-JIMÉNEZ J, ZÚÑIGA G, VILLA-TANACA L, et al. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae Microbial Ecology, 2009, 58(4): 879−891. [9] SHARON G, SEGAL D, RINGO J M, et al. Correction for bencivenni et al., direct asymmetric vinylogous michael addition of cyclic enones to nitroalkenes via dienamine catalysis [J]. Proceedings of the National Academy of Sciences, 2013, 110(12): 4852−4852. [10] VISÔTTO L E, OLIVEIRA M G A, GUEDES R N C, et al. Contribution of gut bacteria to digestion and development of the velvetbean caterpillar, Anticarsia gemmatalis [J]. Journal of Insect Physiology, 2009, 55(3): 185−191. doi: 10.1016/j.jinsphys.2008.10.017 [11] ZHOU F Y, WANG Z, XUE M, et al. Toxicity of selected insecticides for onion maggot Delia antiqua (Meigen) adult and their bait-allure efficacy in the fields [J]. Journal of Food Agriculture and Environment, 2013, 11(3): 1409−1413. [12] THAKUR A, DHAMMI P, SAINI H S, et al. Pathogenicity of bacteria isolated from gut of Spodoptera litura (Lepidoptera: Noctuidae) and fitness costs of insect associated with consumption of bacteria [J]. Journal of Invertebrate Pathology, 2015, 127: 38−46. doi: 10.1016/j.jip.2015.02.007 [13] XU L T, LOU Q Z, CHENG C H, et al. Gut-associated bacteria of Dendroctonus valens and their involvement in verbenone production [J]. Microbial Ecology, 2015, 70(4): 1012−1023. doi: 10.1007/s00248-015-0625-4 [14] 陈娟. 两种木食性白蚁肠道内共生细菌多样性的比较研究 [D]. 武汉: 华中师范大学, 2011.CHEN J. Comparative studies on the diversity of intestinal symbiotic bacteria between two different wood-feeding termites [D]. Wuhan: Central China Normal University, 2011. [15] 张义强. 蜜蜂肠道共生菌的研究 [D]. 福州: 福建农林大学, 2013.ZHANG Y Q. The study of the symbiotic bacteria in honeybee intestinal tract [D]. Fuzhou: Fujian Agriculture and Forestry University, 2013. [16] ADAMS A S, BOONE C K, BOHLMANN J, et al. Responses of bark beetle-associated bacteria to host monoterpenes and their relationship to insect life histories [J]. Journal of Chemical Ecology, 2011, 37(8): 808−817. doi: 10.1007/s10886-011-9992-6 [17] 朱临. 淡色库蚊肠道细菌多样性分析 [D]. 淮南: 安徽理工大学, 2011.ZHU L. The analysis of diversity of bacteria of the Culex Pipiens Pallens' intestinal canal [D]. Huainan: AnHui University of Science and Technology, 2011. (in Chinese) [18] 孙博通, 蓝波妙, 王倩, 等. 斜纹夜蛾幼虫肠道细菌分离鉴定及其功能初步分析 [J]. 生物资源, 2017, 39(4):264−271.SUN B T, LAN M B, WANG Q, et al. lsolation and preliminary functional analysis of the larval gut bacteria from Spodoptera litura [J]. Biotic Resources, 2017, 39(4): 264−271.(in Chinese) [19] 刘玉升, 李明立, 刘俊展, 等. 东亚飞蝗肠道细菌的研究 [J]. 中国微生态学杂志, 2007, 19(1):34−36,39. doi: 10.3969/j.issn.1005-376X.2007.01.014LIU Y S, LI M L, LIU J Z, et al. Study on the intestinal bacteria in L. migratoria manilensis [J]. Chinese Journal of Microecology, 2007, 19(1): 34−36,39.(in Chinese) doi: 10.3969/j.issn.1005-376X.2007.01.014 [20] 张云霞, 薛明, 宋增明. 葱蝇Delia-antiqua(Meigen)的研究进展 [J]. 山东农业大学学报(自然科学版), 2003, 34(3):455−458.ZHANG Y X, XUE M, SONG Z M. Progesses in Delia antiqua(Meigen) study [J]. Journal of Shandong Agricultural University (Natural Science), 2003, 34(3): 455−458.(in Chinese) [21] EYMANN M, FRIEND W G. Development of onion maggots (Diptera: Anthomyiidae) on bacteria-free onion agar supplemented with vitamins and amino acids [J]. Annals of the Entomological Society of America, 1985, 78(2): 182−185. doi: 10.1093/aesa/78.2.182 [22] SCHNEIDER W D, MILLER J R, BREZNAK J A, et al. Onion maggot, Delia antiqua, survival and development on onions in the presence and absence of microorganisms [J]. Entomologia Experimentalis et Applicata, 2011, 33(1): 50−56. [23] SHAO Y Q, CHEN B S, SUN C, et al. Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota [J]. Cell Chemical Biology, 2017, 24(1): 66−75. doi: 10.1016/j.chembiol.2016.11.015 [24] ZHOU F Y, WU X Q, XU L T, et al. Repressed Beauveria bassiana infections in Delia antiqua due to associated microbiota [J]. Pest Management Science, 2019, 75(1): 170−179. doi: 10.1002/ps.5084 [25] ZHANG J Y, LIU Y X, GUO X X, et al. High-throughput cultivation and identification of bacteria from the plant root microbiota [J]. Nature Protocols, 2021, 16(2): 988−1012. doi: 10.1038/s41596-020-00444-7 [26] CAPORASO J G, KUCZYNSKI J, STOMBAUGH J, et al. QIIME allows analysis of high-throughput community sequencing data [J]. Nature Methods, 2010, 7(5): 335−336. doi: 10.1038/nmeth.f.303 [27] EDGAR R C. Search and clustering orders of magnitude faster than BLAST [J]. Bioinformatics, 2010, 26(19): 2460−2461. doi: 10.1093/bioinformatics/btq461 [28] WANG Q, GARRITY G M, TIEDJE J M, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy [J]. Applied and Environmental Microbiology, 2007, 73(16): 5261−5267. doi: 10.1128/AEM.00062-07 [29] QUAST C, PRUESSE E, YILMAZ P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools[J]. Nucleic Acids Research, 2013, 41(Database issue): D590-D596. [30] LIU M, ZHAO X Y, LI X X, et al. Antagonistic effects of Delia antiqua (Diptera: Anthomyiidae)-associated bacteria against four phytopathogens [J]. Journal of Economic Entomology, 2021, 114(2): 597−610. doi: 10.1093/jee/toab002 [31] 王媛媛. 抗镰刀菌芽孢杆菌的分离、筛选、鉴定及其抗菌机理初步研究 [D]. 保定: 河北农业大学, 2013.WANG Y Y. Isolation, screening and identification of anti-Fusarium Bacillus and preliminary study of the disease-suppressive mechanisms. [D]. Baoding: Agriculture University of Hebei, 2013. [32] 庞发虎. 小麦内生细菌的种群多样性调查及其在小麦条锈病生物防治中的利用研究 [D]. 南宁: 广西大学, 2016.PANG F H. Studies on the population diversity of bacterial endophytes in wheat plants and their application in the biocontrol of wheat stripe rust [D]. Nanning: Guangxi University, 2016. (in Chinese) [33] CUI L X, YANG C D, WEI L J, et al. Isolation and identification of an endophytic bacteria Bacillus velezensis 8-4 exhibiting biocontrol activity against potato scab [J]. Biological Control, 2020, 141: 104156. doi: 10.1016/j.biocontrol.2019.104156 [34] 刘婧, 陈丹, 庄桂芬, 等. 家蝇发育过程中肠道可培养共生细菌的分离与鉴定 [J]. 中国寄生虫学与寄生虫病杂志, 2017, 35(2):120−124.LIU J, CHEN D, ZHUANG G F, et al. Isolation and identification of cultivable symbiotic bacteria from the intestinal tract of Musca domestica during development [J]. Chinese Journal of Parasitology and Parasitic Diseases, 2017, 35(2): 120−124.(in Chinese) [35] LIU Y P, XU L T, ZHANG Z Q, et al. Isolation, identification, and analysis of potential functions of culturable bacteria associated with an invasive gall wasp, Leptocybe invasa [J]. Microbial Ecology, 2021: 1−16. [36] LUO A R, ZHANG Y Z, QIAO H J, et al. Outgroup selection in tree reconstruction: a case study of the family Halictidae (Hymenoptera: Apoidea) [J]. Acta Entomologica Sinica, 2010, 53(2): 192−201. [37] LYSYK T J, KALISCHUK-TYMENSEN L, SELINGER L B, et al. Rearing stable fly larvae (Diptera: Muscidae) on an egg yolk medium [J]. Journal of Medical Entomology, 1999, 36(3): 382−388. doi: 10.1093/jmedent/36.3.382 [38] WANG S C, WANG L Y, FAN X, et al. An insight into diversity and functionalities of gut microbiota in insects [J]. Current Microbiology, 2020, 77(9): 1976−1986. doi: 10.1007/s00284-020-02084-2 [39] ENGEL P, MORAN N A. The gut microbiota of insects - diversity in structure and function [J]. FEMS Microbiology Reviews, 2013, 37(5): 699−735. doi: 10.1111/1574-6976.12025 [40] GHODAKE G S, KALME S D, JADHAV J P, et al. Purification and partial characterization of lignin peroxidase from Acinetobacter calcoaceticus NCIM 2890 and its application in decolorization of textile dyes [J]. Applied Biochemistry and Biotechnology, 2009, 152(1): 6−14. doi: 10.1007/s12010-008-8258-4 [41] HU X, LI M, RAFFA K F, et al. Bacterial communities associated with the pine wilt disease vector Monochamus alternatus (Coleoptera: Cerambycidae) during different larval instars [J]. Journal of Insect Science, 2017, 17(6): 115. [42] GUPTA A K, RASTOGI G, NAYDUCH D, et al. Molecular phylogenetic profiling of gut-associated bacteria in larvae and adults of flesh flies [J]. Medical and Veterinary Entomology, 2014, 28(4): 345−354. doi: 10.1111/mve.12054 [43] KSENTINI I, GHARSALLAH H, SAHNOUN M et al. Providencia entomophila sp. nov. , a new bacterial species associated with major olive pests in Tunisia [J]. PLoS One, 2019, 14(10). Doi: 10.1371/journal.pone.0223943 [44] GUJJAR N R, GOVINDAN S, VERGHESE A, et al. Diversity of the cultivable gut bacterial communities associated with the fruit flies Bactrocera dorsalis and Bactrocera cucurbitae (Diptera: Tephritidae) [J]. Phytoparasitica, 2017, 45(4): 453−460. doi: 10.1007/s12600-017-0604-z [45] PANDIARAJAN J, KRISHNAN M. Comparative bacterial survey in the gut of lepidopteran insects with different bionetwork [J]. Microbiology, 2018, 87(1): 103−115. doi: 10.1134/S0026261718010137 [46] PING L Y, BÜCHLER R, MITHÖFER A, et al. A novel Dps-type protein from insect gut bacteria catalyses hydrolysis and synthesis of N-acyl amino acids [J]. Environmental Microbiology, 2007, 9(6): 1572−1583. doi: 10.1111/j.1462-2920.2007.01279.x [47] MEDINA D, WALKE J B, GAJEWSKI Z, et al. Culture media and individual hosts affect the recovery of culturable bacterial diversity from amphibian skin [J]. Frontiers in Microbiology, 2017, 8: 1574. doi: 10.3389/fmicb.2017.01574 [48] FERREIRA F S, HORVATH M B, TONDO E C. Assessing the growth and recovery of Salmonella Enteritidis SE86 after sodium dichloroisocyanurate exposure [J]. Brazilian Journal of Microbiology, 2013, 44(3): 785−790. doi: 10.1590/S1517-83822013000300018 -







 下载:
下载: