Metabolism pathway of malic acid in Saccharomyces cerevisiae JP2 and analysis of its key genes
-
摘要:
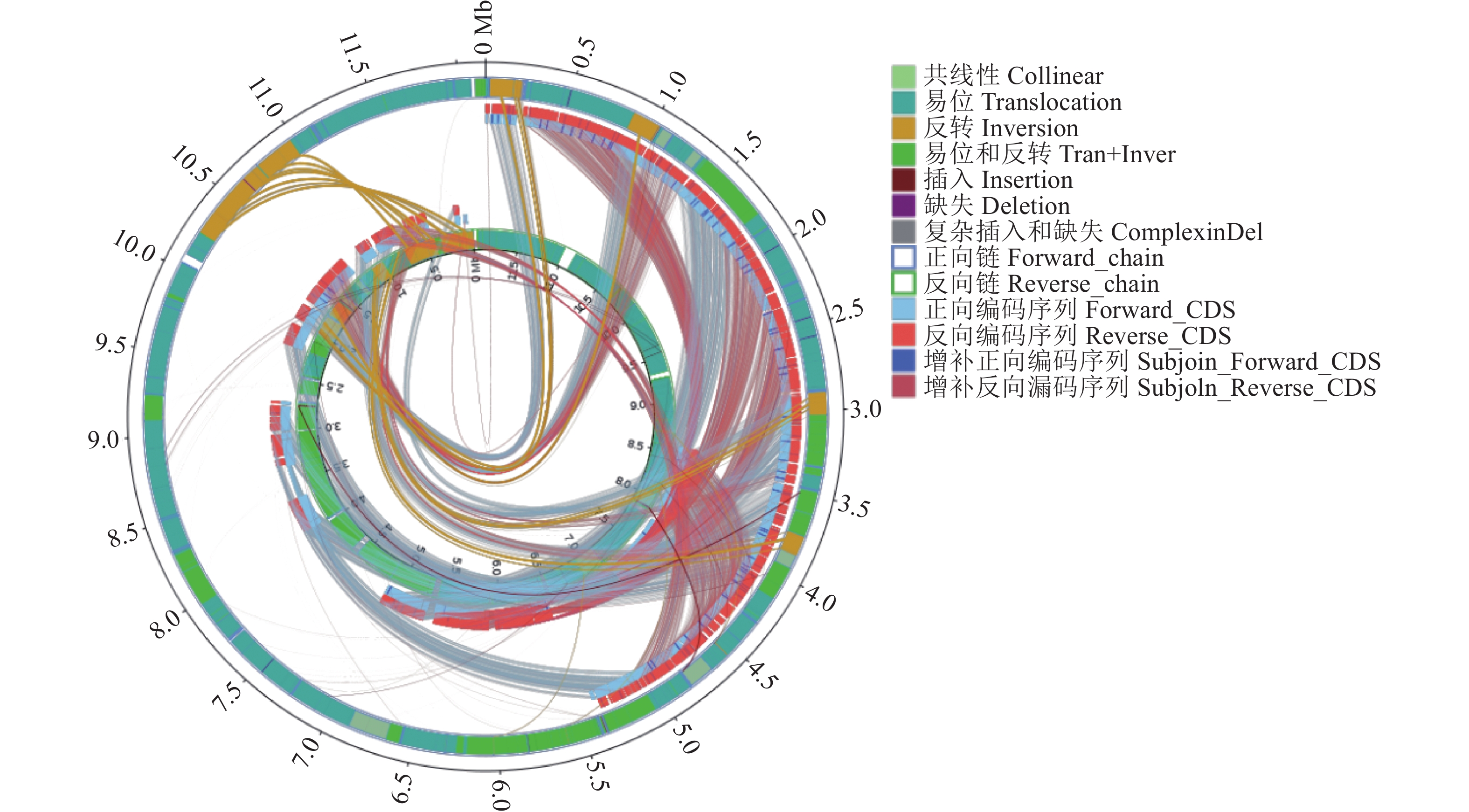
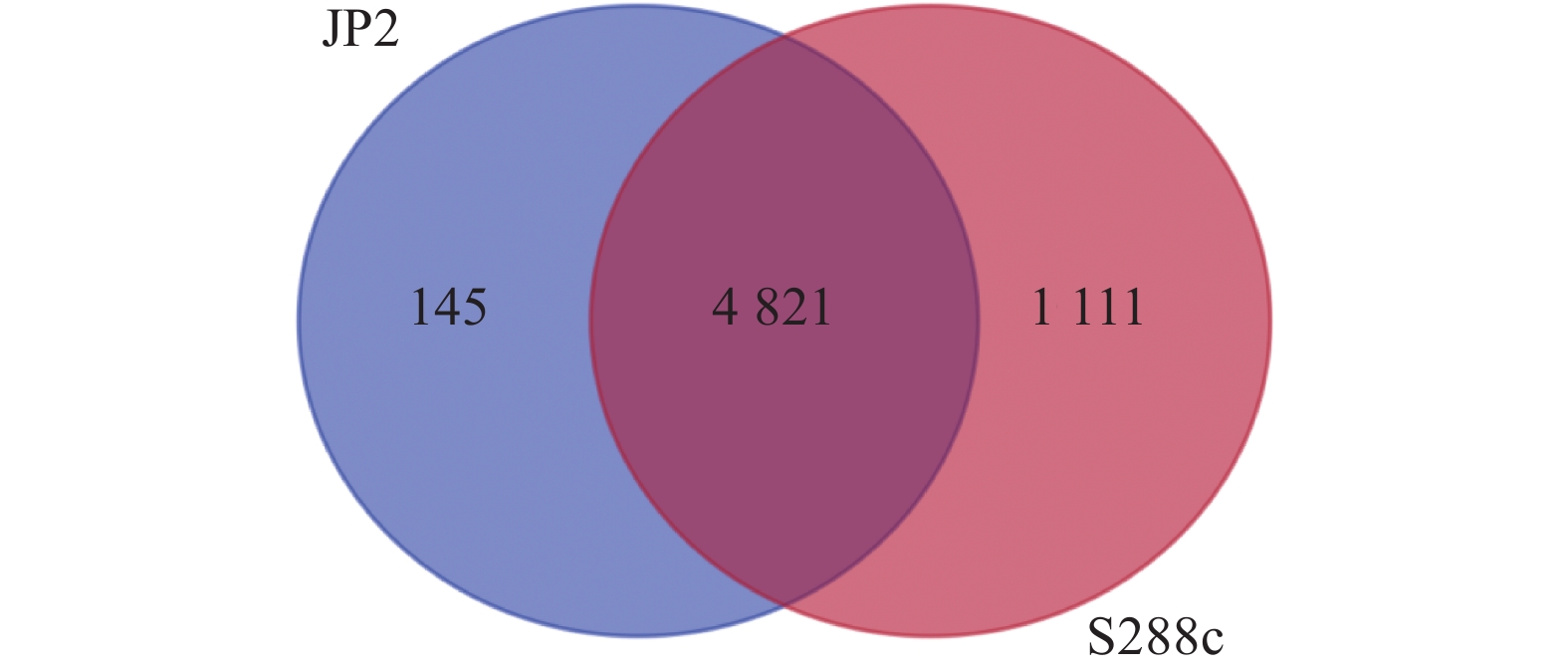
目的 酿酒酵母JP2(Saccharomyces cerevisiae JP2)是从自然发酵枇杷酒醪中分离得到一株具有较好代谢苹果酸的酵母菌。为深入研究其内在降酸机理,对其进行全基因组测序和基因组序列信息解析,并分析其可能的代谢苹果酸途径,找出影响降酸的基因。 方法 基于Illumina Miseq和PacBio测序平台,联用第二代和第三代测序技术对酿酒酵母JP2和模式菌株酿酒酵母S288c进行全基因组测序,采用生物信息学方法对其进行序列组装、基因预测与功能注释,并挖掘JP2可能的苹果酸代谢路径。 结果 (1)通过对酿酒酵母JP2基因组组装共得到52个重叠群,整个基因组大小为11.98 Mbp,GC含量 38.07%;JP2和S288c二者的全基因组大小和GC含量差异不大。(2)JP2代谢苹果酸的途径有3个:苹果酸→草酰乙酸→丙酮酸;苹果酸→丙酮酸;苹果酸→富马酸。相关的主要基因分别为maeA、MDH及fumC,其中苹果酸→草酰乙酸→丙酮酸是主要代谢路径,关键基因是maeA。(3)模拟发酵过程苹果酸代谢及其关键基因表达量变化显示,JP2的苹果酸含量下降了35.8%,下降程度显著高于S288c;与S288c相比,JP2的maeA基因呈现表达极显著上调(P<0.01),在发酵后期,其相对表达量达到起始值的130%。 结论 JP2在果酒发酵过程中呈现maeA基因高效表达,从而显著提高了苹果酸降解效率。研究结果可为酿酒酵母JP2降酸功能基因的研究以及降酸工程菌的研发提供技术支撑。 Abstract:Objective Saccharomyces cerevisiae JP2, isolated from naturally fermented loquat, is a yeast strain with good metabolism of malic acidwine mash. To further investigate its intrinsic acid lowering mechanism, whole genome sequencing and genome sequence information analysis were conducted, and possible metabolic pathways of malic acid were analyzed to identify genes that affect acid lowering. Method Based on the Illumina Miseq and PacBio sequencing platforms, second and third generation sequencing techniques were used to sequence the entire genome of S. cerevisiae JP2 and the type strain S. cerevisiae S288c. Bioinformatics methods were used to assemble their sequences, predict their genes, and annotate their functions, while exploring the possible malic acid metabolism pathways of JP2. Result (1) A total of 52 contigs were obtained by assembling the genome of S. cerevisiae JP2, with a total genome size of 11.98 Mbp and a GC content of 38.07%; There is no significant difference in overall genome size and GC content between JP2 and S288c. (2) There are three pathways for JP2 to metabolize malic acid: malic acid → oxaloacetate → pyruvate; malic acid → pyruvate; and malic acid → fumaric acid. The main genes involved are maeA, MDH, and fumC, among which malic acid → oxaloacetate → pyruvate is the main metabolic pathway with the key gene maeA. (3) The simulated fermentation process showed a decrease of 35.8% in malic acid metabolism and key gene expression levels in JP2, with a significant decrease compared to S288c; Compared with S288c, the maeA gene of JP2 was significantly upregulated (P<0.01), and its relative expression reached 130% of the initial value in the late fermentation stage. Conclusion JP2 exhibits high expression of maeA gene during fruit wine fermentation, significantly improving the efficiency of malic acid degradation. The research results can provide technical support for the study of the acid reducing functional gene of S. cerevisiae JP2 and the development of acid reducing engineering bacteria. -
表 1 基因组基本信息
Table 1. Basic information of genome
特征
Property酿酒酵母JP2
S. cerevisiae JP2酿酒酵母S288c
S. cerevisiae S288c序列总长
Total sequence length/Mbp11.98 12.16 GC含量
GC content/%38.07 38.15 预测基因占比
Predicted gene proportion/%66.50 75.37 预测基因数量
Predicted number of genes5205 5906 预测基因长度
Predicted Gene Length/bp1545 1369 tRNA数量
tRNA quantity289 299 rRNA数量
rRNA quantity4 10 表 2 酿酒酵母JP2和S288c发酵过程有机酸代谢
Table 2. Organic acid metabolism during the fermentation process of S. cerevisiae JP2 and S. cerevisiae S288c (单位:μg·mL−1)
有机酸
Organic acid发酵时间
Fermentation timeJP2试验组
JP2 groupS288c试验组
S288c group草酸 Oxalate 发酵起点 0.00±0.00 0.00±0.00 发酵结束 2.58±0.02 2.76±0.04 酒石酸 Tartaric acid 发酵起点 0.00±0.00 0.00±0.00 发酵结束 6.39±0.05 6.81±0.08 苹果酸 Malic acid 发酵起点 4500.00±0.00 4500.00±0.00 发酵结束 2886.87±150.21 4365.36±143.45* 乳酸 Lactic acid 发酵起点 0.00±0.00 0.00±0.00 发酵结束 44.19±3.14 47.12±4.86 乙酸 Acetic acid 发酵起点 0.00±0.00 0.00±0.00 发酵结束 556.26±25.01 213.14±18.24 富马酸 Fumaric acid 发酵起点 0.00±0.00 0.00±0.00 发酵结束 1.71±0.04 1.98±0.06 *:同一发酵时间不同试验组之间差异显著(P<0.05)。
*: Significant difference between different groups at the same fermentation time (P<0.05).表 3 苹果酸代谢关键基因表达量比较
Table 3. Comparison of expression of key genes of malic acid metabolism
菌株
StrainmaeA基因
maeA geneMDH基因
MDH genefumC基因
fumC geneJP2 1.25** 1.02 0.97 S288C 1.00 1.00 1.00 **:不同菌株之间差异极显著(P<0.01)。
**: Significant difference between different strains (P<0.01). -
[1] GRANDVALET C. Oenococcus oeni: Queen of the cellar, nightmare of geneticists [J]. Microbiology, 2017, 163(3): 297−299. doi: 10.1099/mic.0.000456 [2] JIANG J, SUMBY K M, SUNDSTROM J F, et al. Directed evolution of Oenococcus oeni strains for more efficient malolactic fermentation in a multi-stressor wine environment [J]. Food Microbiology, 2018, 73: 150−159. doi: 10.1016/j.fm.2018.01.005 [3] FERNÁNDEZ-PÉREZ R, TENORIO RODRÍGUEZ C, RUIZ-LARREA F. Fluorescence microscopy to monitor wine malolactic fermentation [J]. Food Chemistry, 2019, 274: 228−233. doi: 10.1016/j.foodchem.2018.08.088 [4] VILELA A. Use of nonconventional yeasts for modulating wine acidity [J]. Fermentation, 2019, 5(1): 27. doi: 10.3390/fermentation5010027 [5] 李华, 刘延琳, 蒋思欣, 等. Oenococcus oeni苹果酸-乳酸发酵关键酶基因在酿酒酵母中的转化与表达 [J]. 农业生物技术学报, 2006, 14(4):606−611.LI H, LIU Y L, JIANG S X, et al. Cloning of two key enzyme genes from Oenococcus oeni for malolactic fermentation and their expressions in Saccharomyces cerevisiae [J]. Journal of Agricultural Biotechnology, 2006, 14(4): 606−611.(in Chinese) [6] BENITO Á, JEFFARES D, PALOMERO F, et al. Selected Schizosaccharomyces pombe strains have characteristics that are beneficial for winemaking [J]. PLoS One, 2016, 11(3): e0151102. doi: 10.1371/journal.pone.0151102 [7] PETRUZZI L, CAPOZZI V, BERBEGAL C, et al. Microbial resources and enological significance: Opportunities and benefits [J]. Frontiers in Microbiology, 2017, 8: 995. doi: 10.3389/fmicb.2017.00995 [8] BENITO S, PALOMERO F, CALDERÓN F, et al. Selection of appropriate Schizosaccharomyces strains for winemaking [J]. Food Microbiology, 2014, 42: 218−224. doi: 10.1016/j.fm.2014.03.014 [9] MYLONA A E, DEL FRESNO J M, PALOMERO F, et al. Use of Schizosaccharomyces strains for wine fermentation—Effect on the wine composition and food safety [J]. International Journal of Food Microbiology, 2016, 232: 63−72. doi: 10.1016/j.ijfoodmicro.2016.05.023 [10] MINNAAR P P, JOLLY N P, PAULSEN V, et al. Schizosaccharomyces pombe and Saccharomyces cerevisiae yeasts in sequential fermentations: Effect on phenolic acids of fermented Kei-apple (Dovyalis caffra L. ) juice [J]. International Journal of Food Microbiology, 2017, 257: 232−237. doi: 10.1016/j.ijfoodmicro.2017.07.004 [11] VAUDANO E, COSTANTINI A, GARCIA-MORUNO E. An event-specific method for the detection and quantification of ML01, a genetically modified Saccharomyces cerevisiae wine strain, using quantitative PCR [J]. International Journal of Food Microbiology, 2016, 234: 15−23. doi: 10.1016/j.ijfoodmicro.2016.06.017 [12] BENITO Á, CALDERÓN F, BENITO S. Schizosaccharomyces pombe biotechnological applications in winemaking [J]. Methods in Molecular Biology, 2018, 1721: 217−226. [13] 何志刚, 李维新, 梁璋成, 等. 优良降酸酿酒酵母的分离和鉴定 [J]. 中国食品学报, 2013, 13(5):191−197. doi: 10.16429/j.1009-7848.2013.05.028HE Z G, LI W X, LIANG Z C, et al. Separation and identification of selected yeast with deacidification ability for brewing in fruit wine [J]. Journal of Chinese Institute of Food Science and Technology, 2013, 13(5): 191−197.(in Chinese) doi: 10.16429/j.1009-7848.2013.05.028 [14] 李维新, 魏巍, 何志刚, 等. 酿酒酵母JP2发酵枇杷汁的有机酸代谢及其产香特征分析 [J]. 中国食品学报, 2016, 16(2):251−257. doi: 10.16429/j.1009-7848.2016.02.034LI W X, WEI W, HE Z G, et al. Analysis of organic acids metabolism and fragrant characteristic for fermentated loquat juice using Saccharomyces cerevisiae JP2 [J]. Journal of Chinese Institute of Food Science and Technology, 2016, 16(2): 251−257.(in Chinese) doi: 10.16429/j.1009-7848.2016.02.034 [15] LIANG Z C, LIN X Z, HE Z G, et al. Amino acid and microbial community dynamics during the fermentation of Hong Qu glutinous rice wine [J]. Food Microbiology, 2020, 90: 103467. doi: 10.1016/j.fm.2020.103467 [16] COMITINI F, AGARBATI A, CANONICO L, et al. Yeast interactions and molecular mechanisms in wine fermentation: A comprehensive review [J]. International Journal of Molecular Sciences, 2021, 22(14): 7754. doi: 10.3390/ijms22147754 [17] DARBANI B, STOVICEK V, VAN DER HOEK S A, et al. Engineering energetically efficient transport of dicarboxylic acids in yeast Saccharomyces cerevisiae [J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(39): 19415−19420. doi: 10.1073/pnas.1900287116 [18] BOVO B, NADAI C, VENDRAMINI C, et al. Aptitude of Saccharomyces yeasts to ferment unripe grapes harvested during cluster thinning for reducing alcohol content of wine [J]. International Journal of Food Microbiology, 2016, 236: 56−64. doi: 10.1016/j.ijfoodmicro.2016.07.022 -








 下载:
下载: