Bioinformatics and Expression of CaTPS9 in Chili Peppers
-
摘要:
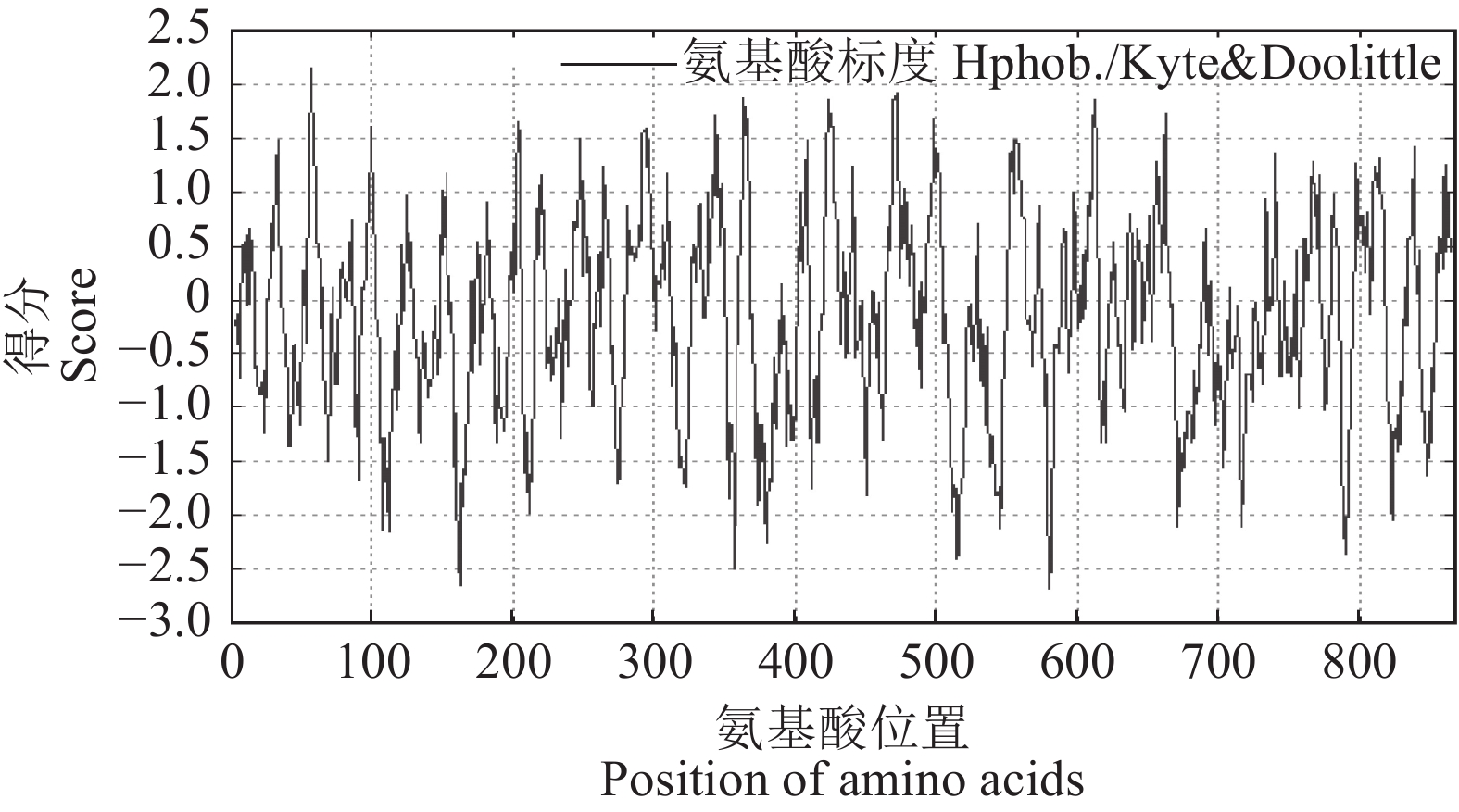
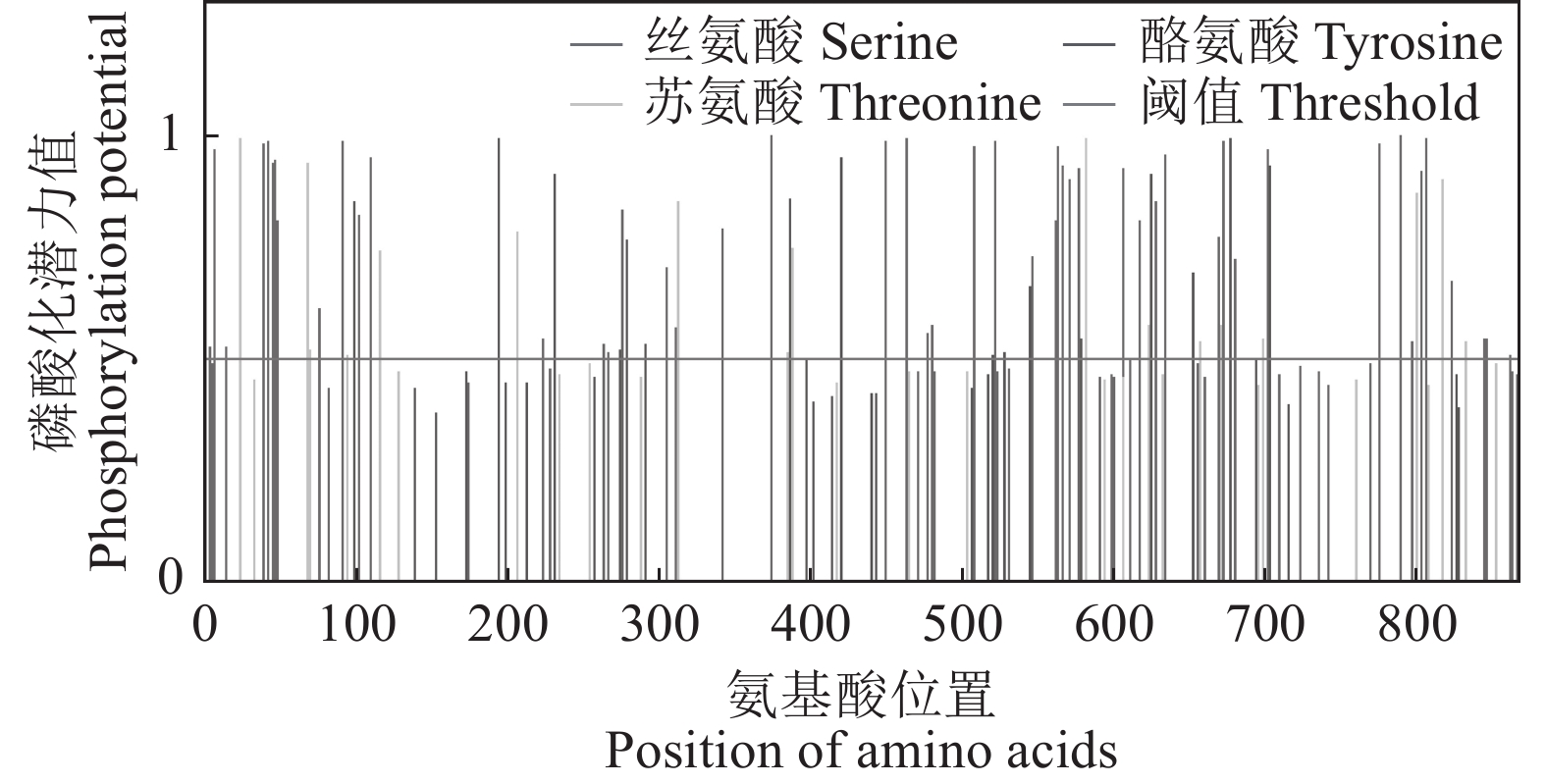
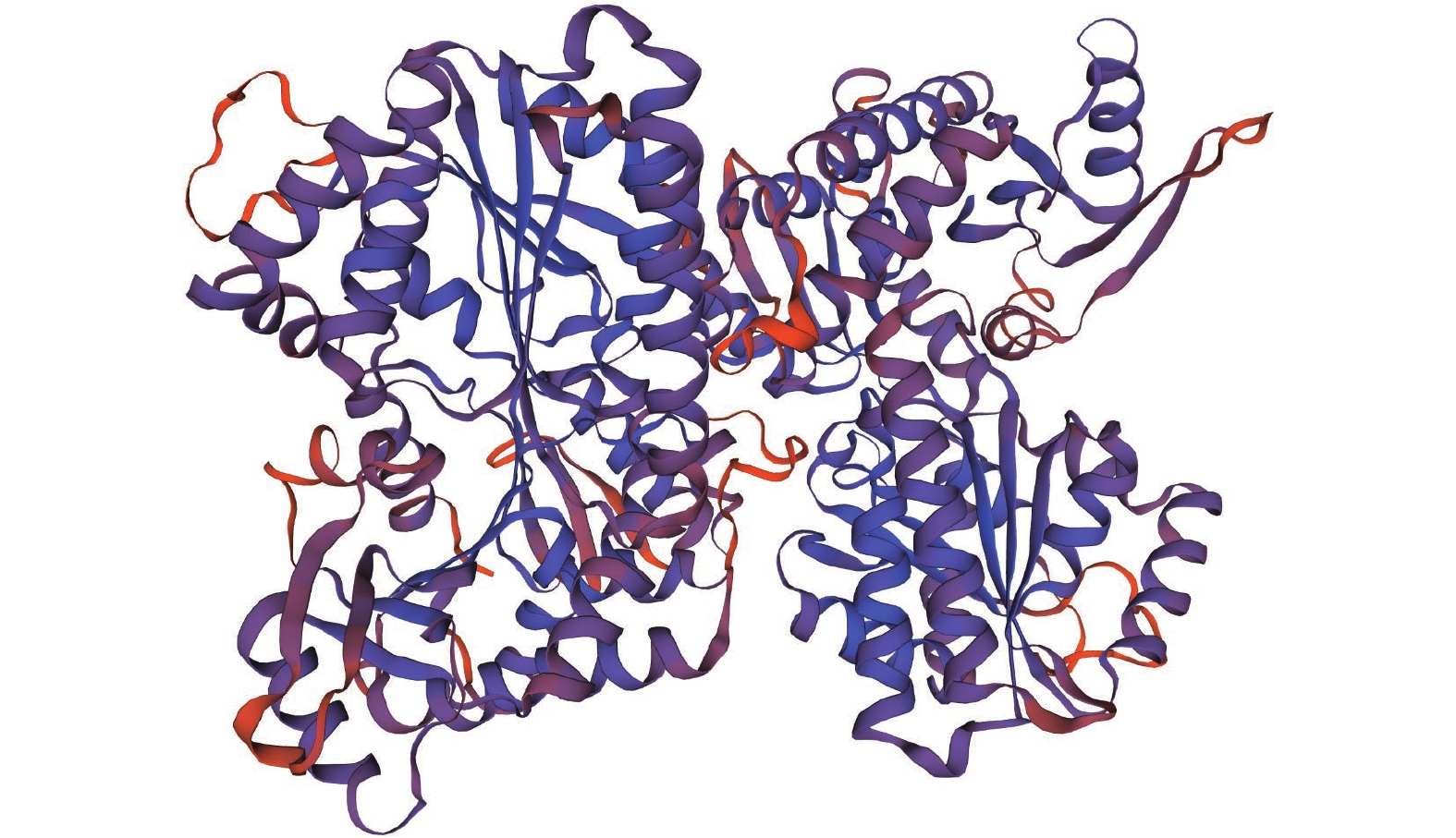
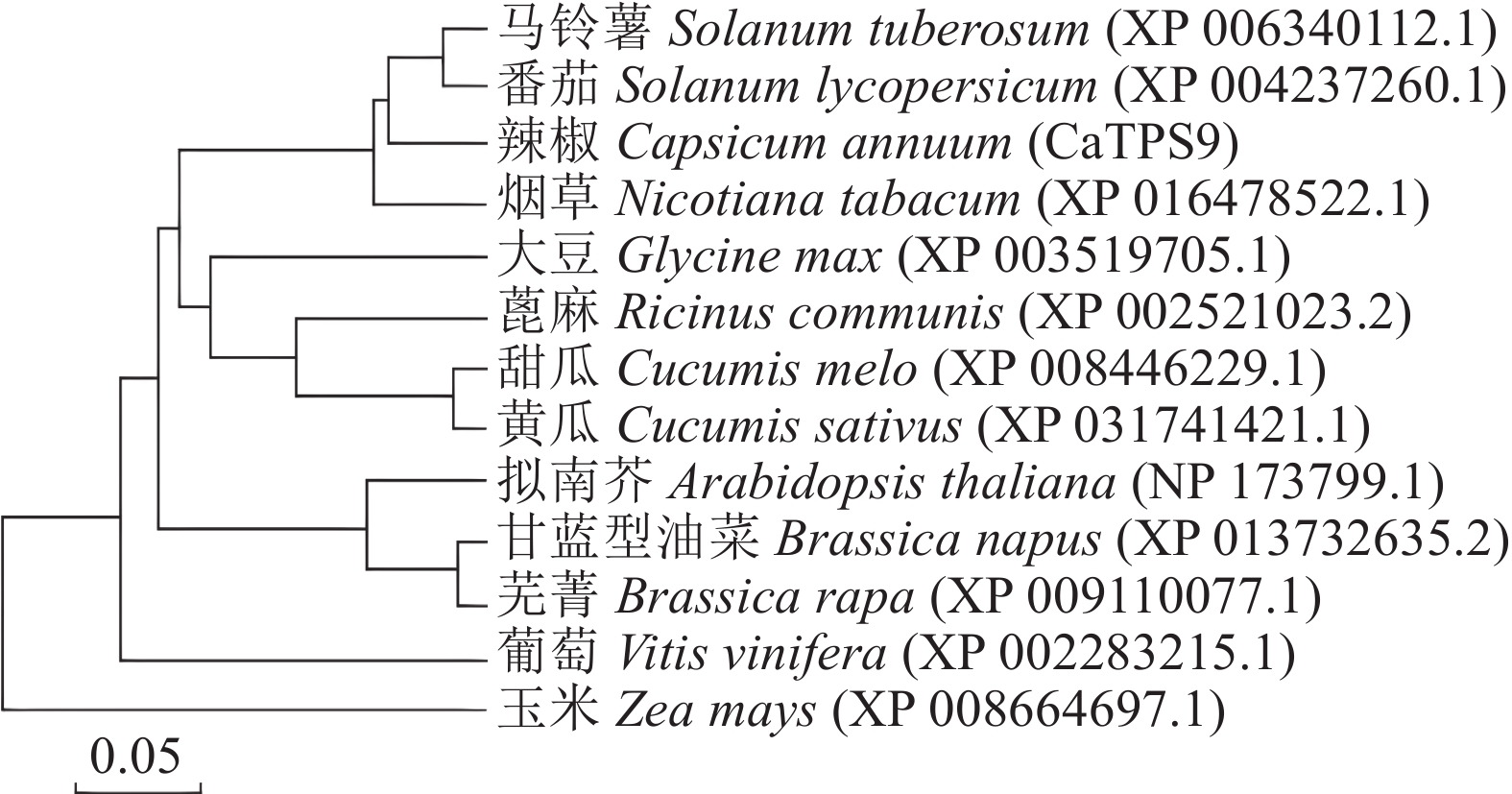
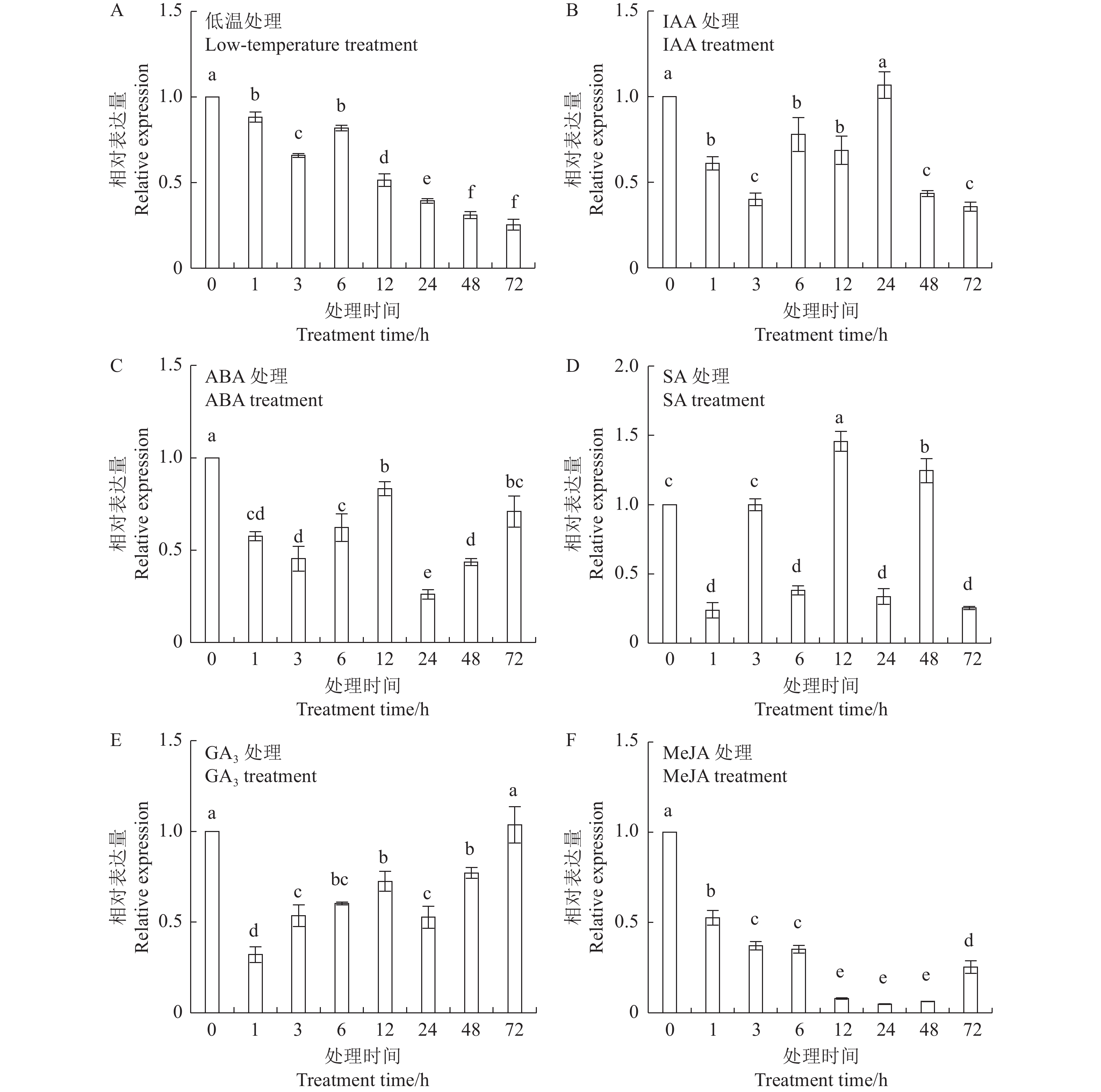
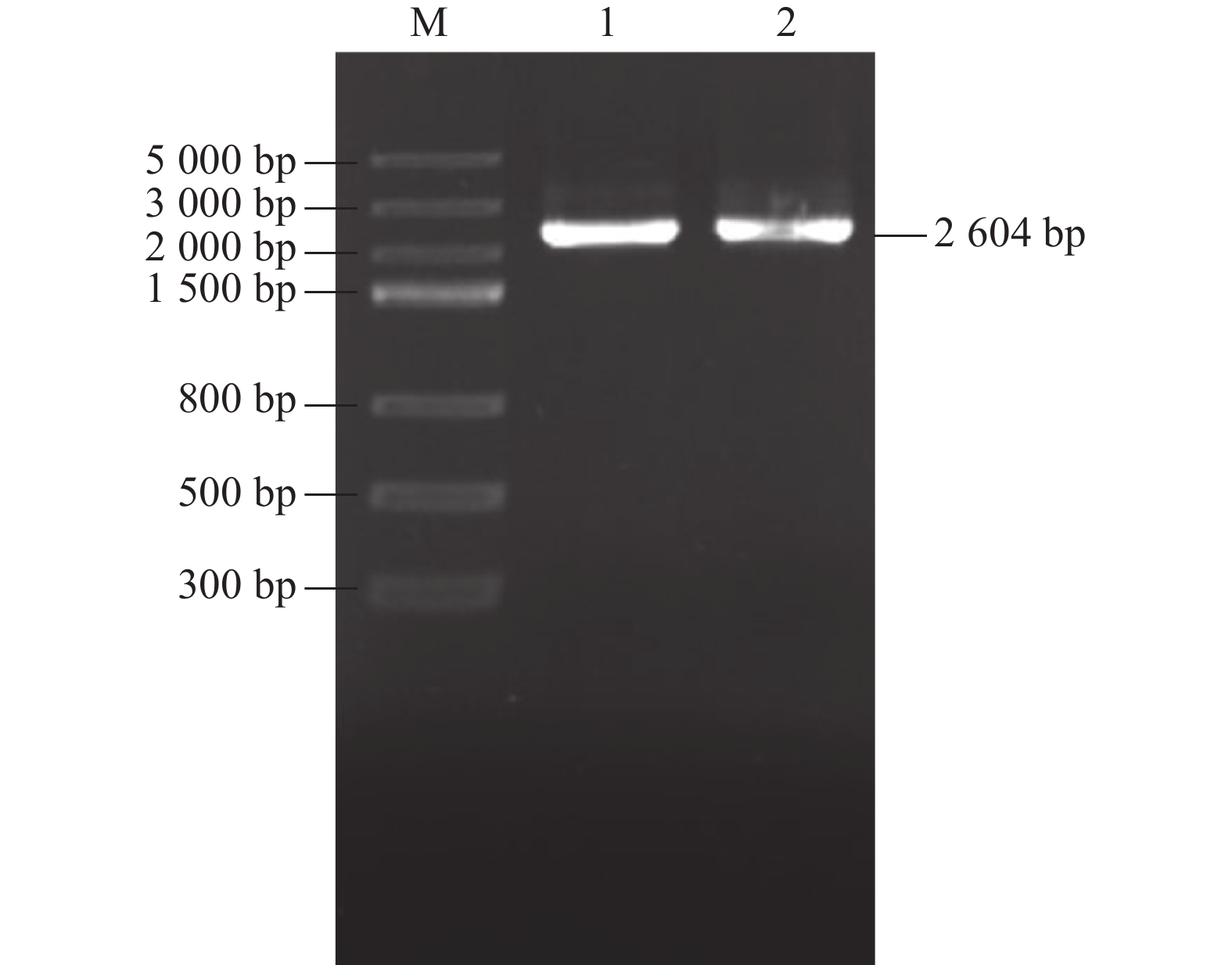
目的 明确辣椒中海藻糖-6-磷酸合酶(TPS)基因CaTPS9的表达特性和生物学功能,进一步了解TPS对辣椒生长中调控非生物胁迫的作用。 方法 以辣椒品种强丰101为试验材料,克隆CaTPS9基因,并对辣椒CaTPS9的理化性质、蛋白结构、顺式作用元件、系统进化树等进行分析;通过qRT-PCR分析CaTPS9基因在不同组织(商品果果肉、幼果果肉、成熟果果肉、商品果胎座、幼果胎座、成熟果胎座、叶、根、茎、花)和胁迫处理(低温和植物生长调节剂处理)中的表达模式。 结果 CaTPS9基因CDS序列全长2604 bp,编码867个氨基酸。CaTPS9蛋白包含Glyco_transf_20和Trehalose_PPase两个保守结构域,分子质量为97.60 kDa,不稳定指数为44.27,理论等电点为5.63,亚细胞定位预测CaTPS9蛋白位于细胞质中。生物信息学分析表明,CaTPS9蛋白属于亲水性蛋白,且不存在跨膜结构和信号肽序列,蛋白结构主要由α-螺旋和无规则卷曲组成。系统进化关系分析表明,CaTPS9与烟草(Nicotiana tabacum L.)、番茄(Solanum lycopersicum L.)和马铃薯(Solanum tuberosum L.)中的同源基因亲缘关系较近。启动子顺式作用元件分析表明,CaTPS9启动子区域含有与激素、胁迫及植物生长发育相关的顺式作用元件。此外,CaTPS9在叶片中表达量最高,在商品果胎座中表达量最低。在水杨酸(Salicylic acid,SA) 处理12 h后CaTPS9基因的表达量被显著提升,而低温、吲哚乙酸(3-indoleacetic acid,IAA)、脱落酸(Abscisic acid,ABA)、赤霉素(Gibberellin acid,GA3)和茉莉酸甲酯(Methyl jasmonate,MeJA)处理能够显著抑制CaTPS9基因表达量。 结论 CaTPS9基因可能通过海藻糖生物合成途径响应逆境胁迫。 -
关键词:
- 辣椒 /
- 海藻糖-6-磷酸合酶 /
- 生物信息学 /
- 非生物胁迫 /
- 表达分析
Abstract:Objective Bioinformatics and expression of trehalose-6-phosphate synthase (TPS) gene in chili peppers,CaTPS9, were studied to understand its role in response to abiotic stress during the plant growth. Method CaTPS9 was cloned from Qiangfeng 101 to analyze the physicochemical properties, protein structure, cis-regulatory element, and phylogenetic tree. Expressions of the gene in the pericarp and placenta of a commercial pepper as well as the young pericarp, ripened pericarp, young placenta, ripened placenta, leaves, roots, stems, and flowers of chili pepper plants under varied stresses including low temperature, 3-indoleacetic acid(IAA), abscisic acid(ABA), salicylic acid(SA), gibberellin A3(GA3), and methyl jasmonate(MeJA) were determined using qRT PCR. Result The full-length cDNA sequence of CaTPS9 was 2604 bp encoded 867 amino acids. Located in the cytoplasm, the protein contained two conserved domains, Glyco_transf_20 and Trehalose_PPase, with a molecular weight of 97.60 kDa, an instability index of 44.27, and a theoretical isoelectric point of 5.63. It was a hydrophilic protein free of transmembrane structure and signal peptide sequence with a structure consisting mainly of alpha helixes and random coils. Phylogenetically, CaTPS9 was close to the homologous genes in Nicotiana tabacum, Solanum lycopersicum and Solanum tuberosum. It had hormones response, stress response, and plant growth and development cis−elements and was expressed most highly in the leaves, lowly in the placentas of a commercially available pepper, and significantly by 12 h SA induction. Stress, such as low temperature, IAA, ABA, GA3, and MeJA, significantly inhibited the expression. Conclusion It appeared that CaTPS9 might regulate the response of chili pepper plants to various abiotic stresses through the trehalose biosynthetic pathway. -
Key words:
- Chili pepper /
- TPS /
- bioinformatics /
- abiotic stress /
- expression analysis
-
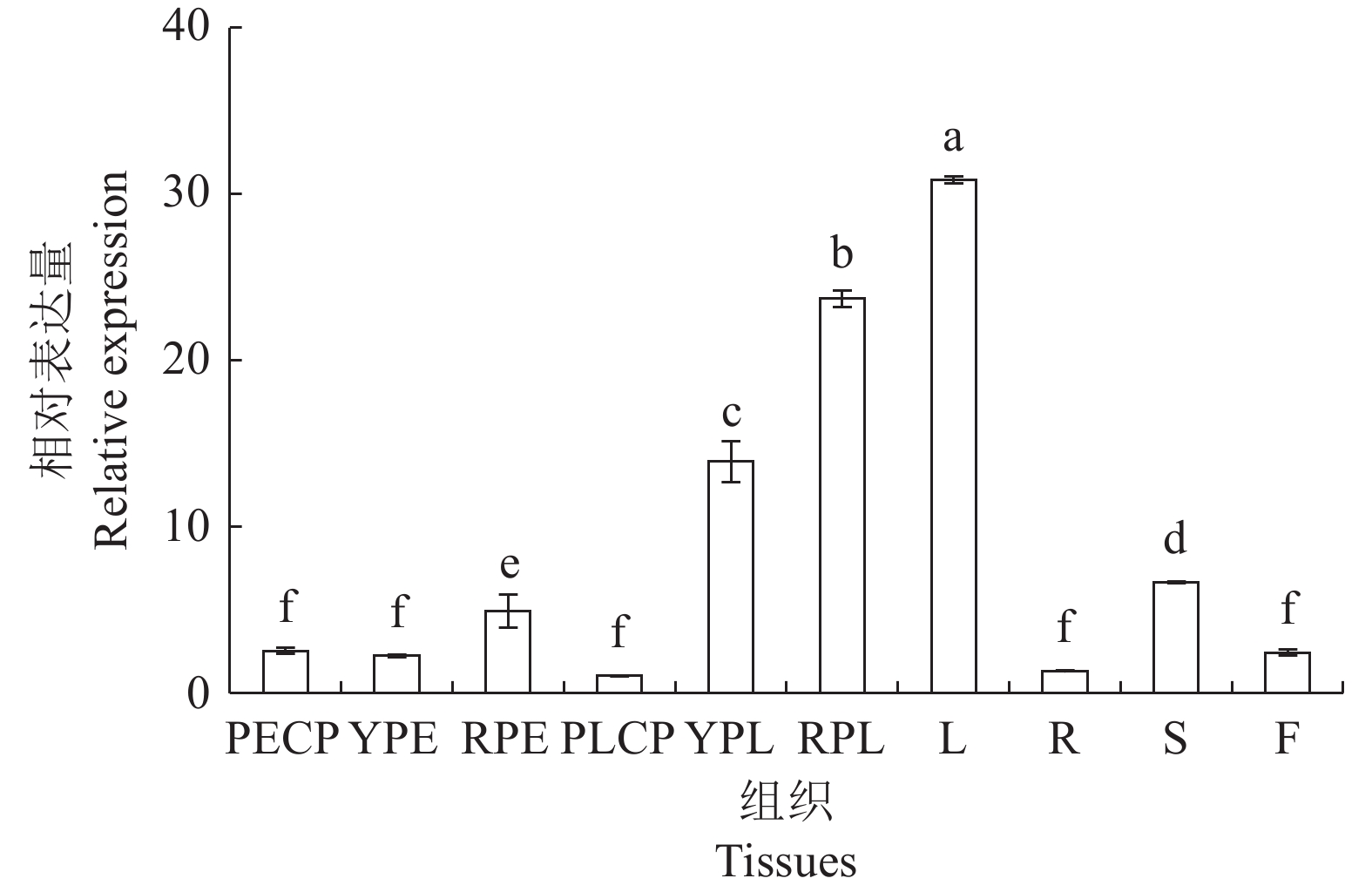
图 7 CaTPS9基因在辣椒不同组织的表达模式
PECP:商品果果肉;YPE:幼果果肉;RPE:成熟果果肉;PLCP:商品果胎座;YPL:幼果胎座;RPL:成熟果胎座;L:叶;R:根;S:茎;F:花;不同小写字母表示CaTPS9基因表达量在辣椒不同组织间差异显著(P<0.05)。图8同。
Figure 7. Expressions of CaTPS9 in different tissues of pepper plant
PECP: pericarp of commerical pepper; YPE: young pericarp; RPE: ripened pericarp; PLCP: placenta of commercial pepper; YPL: young placenta; RPL: ripened placenta ; L: leaf; R: root; S: stem; F: flower; those with different lowercase letters significantly different between different tissues of pepper at P<0.05. Same for Fig. 8.
表 1 CaTPS9基因引物信息及功能
Table 1. Information and function of CaTPS9 primer
引物名称
Primer name引物序列
Primer sequence(5′-3′)用途
PurposeCaTPS9-F ATGGCATCAAGATCTAGTGCA 基因扩增
Gene amplificationCaTPS9-R TTACCCACTCAAATTAACAGATGAG qCaTPS9-F GCATTGGAGATGACAGGTCGGATG 荧光定量PCR
qRT-PCRqCaTPS9-R ACTTGGCTTTGCTTGGCTTTTGC qActin-F AGAGATTCCGTTGCCCAGAGGTC 内参基因
Reference geneqActin-R AGCCACCACTGAGCACAATGTTAC 表 2 CaTPS9启动子顺式作用元件预测分析
Table 2. Putative cis−element analysis on promoter regions of CaTPS9
分类
Classification元件名称
Element name序列
Sequence功能预测
Function prediction激素响应元件
Hormone response element脱落酸响应元件
ABREACGTG 参与响应脱落酸的顺式作用元件
cis-acting element involved in abscisic acid responsiveness生长素响应元件
AuxRR-coreGGTCCAT 参与响应生长素的顺式作用元件
cis-acting regulatory element involved in auxin responsiveness赤霉素响应元件
TATC-boxTATCCCA 参与响应赤霉素的顺式作用元件
cis-acting element involved in gibberellin-responsiveness水杨酸响应元件
TCA-elementCCATCTTTTT 参与响应水杨酸的顺式作用元件
cis-acting element involved in salicylic acid responsiveness胁迫响应元件
Stress response element干旱诱导元件
MBSCAACTG 参与干旱诱导的MYB结合位点
MYB binding site involved in drought-inducibility厌氧诱导元件
AREAAACCA 参与厌氧诱导的顺式作用元件
cis-acting regulatory element essential for the anaerobic induction植物生长发育元件
Plant growth and development elements玉米蛋白代谢调节元件
O2-siteGATGA(C/T)
(A/G)TG(A/G)参与玉米蛋白代谢调节的顺式作用元件
cis-acting regulatory element involved in zein metabolism regulation种子特异调控元件
RY-elementCATGCATG 参与种子特异调控的顺式作用元件
cis-acting regulatory element involved in seed-specific regulation分生组织表达相关元件
CAT-boxGCCACT 与分生组织表达相关的顺式作用元件
cis-acting regulatory element related to meristem expression -
[1] 王立浩, 马艳青, 张宝玺. 我国辣椒品种市场需求与育种趋势 [J]. 中国蔬菜, 2019(8):1−4. doi: 10.19928/j.cnki.1000-6346.2019.08.001WANG L H, MA Y Q, ZHANG B X. Market demand and breeding trend of pepper varieties in China [J]. China Vegetables, 2019(8): 1−4.(in Chinese) doi: 10.19928/j.cnki.1000-6346.2019.08.001 [2] ZHU F M, LI M Y, SUN M L, et al. Plant hormone signals regulate trehalose accumulation against osmotic stress in watermelon cells [J]. Protoplasma, 2022, 259(5): 1351−1369. doi: 10.1007/s00709-021-01715-0 [3] GARG A K, KIM J K, OWENS T G, et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses [J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(25): 15898−15903. [4] SINGER M A, LINDQUIST S. Multiple effects of trehalose on protein folding in vitro and in vivo [J]. Molecular Cell, 1998, 1(5): 639−648. doi: 10.1016/S1097-2765(00)80064-7 [5] CHOWDARY T K, RAMAN B, RAMAKRISHNA T, et al. Interaction of mammalian Hsp22 with lipid membranes [J]. The Biochemical Journal, 2007, 401(2): 437−445. doi: 10.1042/BJ20061046 [6] GARCIA A B, ENGLER J, IYER S, et al. Effects of osmoprotectants upon NaCl stress in rice [J]. Plant Physiology, 1997, 115(1): 159−169. doi: 10.1104/pp.115.1.159 [7] TIAN L F, XIE Z J, LU C Q, et al. The trehalose-6-phosphate synthase TPS5 negatively regulates ABA signaling in Arabidopsis thaliana [J]. Plant Cell Reports, 2019, 38(8): 869−882. doi: 10.1007/s00299-019-02408-y [8] 周斌辉. 巴西橡胶树6-磷酸海藻糖合成酶家族基因的克隆、表达分析及其功能验证[D]. 海口: 海南大学, 2013.ZHOU B H. Cloning and functional characterization of the trehalose-6-phosphate synthase gene family in Hevea brasiliensis[D]. Haikou: Hainan University, 2013. (in Chinese) [9] LI H W, ZANG B S, DENG X W, et al. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice [J]. Planta, 2011, 234(5): 1007−1018. doi: 10.1007/s00425-011-1458-0 [10] WAHL V, PONNU J, SCHLERETH A, et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana [J]. Science, 2013, 339(6120): 704−707. doi: 10.1126/science.1230406 [11] VANDESTEENE L, RAMON M, LE ROY K, et al. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis [J]. Molecular Plant, 2010, 3(2): 406−419. doi: 10.1093/mp/ssp114 [12] ZANG B S, LI H W, LI W J, et al. Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice [J]. Plant Molecular Biology, 2011, 76(6): 507−522. doi: 10.1007/s11103-011-9781-1 [13] 杨仕梅, 张天缘, 丘日光, 等. 番茄TPS基因家族鉴定与分析 [J]. 分子植物育种, 2019, 17(16):5215−5223.YANG S M, ZHANG T Y, QIU R G, et al. Identification and analysis of TPS gene family in tomato [J]. Molecular Plant Breeding, 2019, 17(16): 5215−5223.(in Chinese) [14] 魏兵强, 王兰兰, 张茹, 等. 辣椒TPS家族成员的鉴定与CaTPS1的表达分析 [J]. 园艺学报, 2016, 43(8):1504−1512.WEI B Q, WANG L L, ZHANG R, et al. Identification of CaTPS gene family and expression analysis of CaTPS1 in hot pepper [J]. Acta Horticulturae Sinica, 2016, 43(8): 1504−1512.(in Chinese) [15] 陈天池, 吴月燕, 沈乐意, 等. 葡萄TPS基因家族的鉴定与表达分析[J/OL]. 分子植物育种, 2021: 1-13. (2021-07-22). https://kns.cnki.net/kcms/detail/46.1068.S.20210722.1430.018.html.CHEN T C, WU Y Y, SHEN L Y, et al. Identification and expression analysis of TPS genes family in grape[J/OL]. Molecular Plant Breeding, 2021: 1-13. (2021-07-22). https://kns.cnki.net/kcms/detail/46.1068.S.20210722.1430.018.html.(in Chinese) [16] 杜丽璞, 徐惠君, 叶兴国, 等. 小麦转TPS基因植株的获得及其初步功能鉴定 [J]. 麦类作物学报, 2007, 27(3):369−373.DU L P, XU H J, YE X G, et al. Transgenic wheat plants with trehalose-6-phosphate synthase (TPS) gene and identification of their function [J]. Journal of Triticeae Crops, 2007, 27(3): 369−373.(in Chinese) [17] CORTINA C, CULIÁÑEZ-MACIÀ F A. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis [J]. Plant Science, 2005, 169(1): 75−82. doi: 10.1016/j.plantsci.2005.02.026 [18] 赵淑芳, 苟秉调, 魏敏, 等. 辣椒CaTPS8基因克隆与表达分析 [J]. 西北农业学报, 2022, 31(12):1568−1578.ZHAO S F, GOU B D, WEI M, et al. Cloning and expression analysis of CaTPS8 gene in Capsicum annuum [J]. Acta Agriculturae Boreali-Occidentalis Sinica, 2022, 31(12): 1568−1578.(in Chinese) [19] LU S N, WANG J Y, CHITSAZ F, et al. CDD/SPARCLE: The conserved domain database in 2020 [J]. Nucleic Acids Research, 2020, 48(D1): D265−D268. doi: 10.1093/nar/gkz991 [20] WILKINS M R, GASTEIGER E, BAIROCH A, et al. Protein identification and analysis tools in the ExPASy server [J]. Methods in Molecular Biology, 1999, 112: 531−552. [21] CHOU K C, SHEN H B. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0 [J]. PLoS One, 2010, 5(4): e9931. doi: 10.1371/journal.pone.0009931 [22] JOHNSON M, ZARETSKAYA I, RAYTSELIS Y, et al. NCBI BLAST: A better web interface [J]. Nucleic Acids Research, 2008, 36(S2): W5−W9. [23] LARKIN M A, BLACKSHIELDS G, BROWN N P, et al. Clustal W and clustal X version 2.0 [J]. Bioinformatics, 2007, 23(21): 2947−2948. doi: 10.1093/bioinformatics/btm404 [24] MÖLLER S, CRONING M D R, APWEILER R. Evaluation of methods for the prediction of membrane spanning regions [J]. Bioinformatics, 2001, 17(7): 646−653. doi: 10.1093/bioinformatics/17.7.646 [25] ALMAGRO A J J, TSIRIGOS K D, SØNDERBY C K, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks [J]. Nature Biotechnology, 2019, 37(4): 420−423. doi: 10.1038/s41587-019-0036-z [26] BLOM N, GAMMELTOFT S, BRUNAK S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites 1 [J]. Journal of Molecular Biology, 1999, 294(5): 1351−1362. doi: 10.1006/jmbi.1999.3310 [27] GEOURJON C, DELÉAGE G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments [J]. Bioinformatics, 1995, 11(6): 681−684. doi: 10.1093/bioinformatics/11.6.681 [28] KIEFER F, ARNOLD K, KÜNZLI M, et al. The SWISS-MODEL Repository and associated resources[J]. Nucleic Acids Research, 2009, 37(Database issue): D387-D392. [29] KUMAR S, STECHER G, LI M, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms [J]. Molecular Biology and Evolution, 2018, 35(6): 1547−1549. doi: 10.1093/molbev/msy096 [30] ROMBAUTS S, DÉHAIS P, VAN MONTAGU M, et al. PlantCARE, a plant cis-acting regulatory element database [J]. Nucleic Acids Research, 1999, 27(1): 295−296. doi: 10.1093/nar/27.1.295 [31] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method [J]. Methods, 2001, 25(4): 402−408. doi: 10.1006/meth.2001.1262 [32] JIANG S Y, JIN J J, SAROJAM R, et al. A comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns [J]. Genome Biology and Evolution, 2019, 11(8): 2078−2098. doi: 10.1093/gbe/evz142 [33] 于欢. 甘薯海藻糖-6-磷酸合成酶基因IbTPS1克隆与功能鉴定[D]. 太谷: 山西农业大学, 2019.YU H. Cloning and functional identification of trehalose-6-phosphate synthase gene IbTPS1 from Ipomoea batatas(L. ) lam[D]. Taigu: Shanxi Agricultural University, 2019. (in Chinese) [34] 丁泽红, 付莉莉, 铁韦韦, 等. 木薯MeTPS9基因克隆及表达特性分析 [J]. 生物技术通报, 2017, 33(11):84−91. doi: 10.13560/j.cnki.biotech.bull.1985.2017-0416DING Z H, FU L L, TIE W W, et al. Clone and expression characteristics of MeTPS9 gene in cassava [J]. Biotechnology Bulletin, 2017, 33(11): 84−91.(in Chinese) doi: 10.13560/j.cnki.biotech.bull.1985.2017-0416 [35] DING X D, WANG D, XIAO J L. Cloning of gs TPS9 gene from Glycine soja and study on its responses to stresses [J]. Journal of Northeast Agricultural University (English Edition), 2022, 29(1): 59−68. [36] KHATUN K, ROBIN A H K, PARK J I, et al. Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones [J]. International Journal of Molecular Sciences, 2017, 18(5): 1056. doi: 10.3390/ijms18051056 [37] DOSSA K, DIOUF D, CISSÉ N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response [J]. Frontiers in Plant Science, 2016, 7: 1522. [38] LIN T Y, ZHOU R, BI B, et al. Analysis of a radiation-induced dwarf mutant of a warm-season turf grass reveals potential mechanisms involved in the dwarfing mutant [J]. Scientific Reports, 2020, 10(1): 18913. doi: 10.1038/s41598-020-75421-x [39] SHINOZAKI K, YAMAGUCHI-SHINOZAKI K. Gene networks involved in drought stress response and tolerance [J]. Journal of Experimental Botany, 2007, 58(2): 221−227. [40] SUGANO S, MAEDA S, HAYASHI N, et al. Tyrosine phosphorylation of a receptor-like cytoplasmic kinase, BSR1, plays a crucial role in resistance to multiple pathogens in rice [J]. The Plant Journal:for Cell and Molecular Biology, 2018, 96(6): 1137−1147. doi: 10.1111/tpj.14093 [41] XU Y C, WANG Y J, MATTSON N, et al. Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: Evolution and differential expression during development and stress [J]. BMC Genomics, 2017, 18(1): 926. doi: 10.1186/s12864-017-4298-x [42] ZHOU M L, ZHANG Q, SUN Z M, et al. Trehalose metabolism-related genes in maize [J]. Journal of Plant Growth Regulation, 2014, 33(2): 256−271. doi: 10.1007/s00344-013-9368-y [43] 龙娅丽, 徐子健, 朱白婢, 等. 茉莉酸甲酯诱导西瓜ClTPS1基因的表达特性及其生物学信息分析 [J]. 分子植物育种, 2016, 14(12):3299−3307.LONG Y L, XU Z J, ZHU B B, et al. Sequence analysis of ClTPS1 gene in watermelon and its expression characteristic after treated with MeJA [J]. Molecular Plant Breeding, 2016, 14(12): 3299−3307.(in Chinese) -







 下载:
下载: