Gene Cloning and Functional Analysis of MebZIP2 in Cassava
-
摘要:
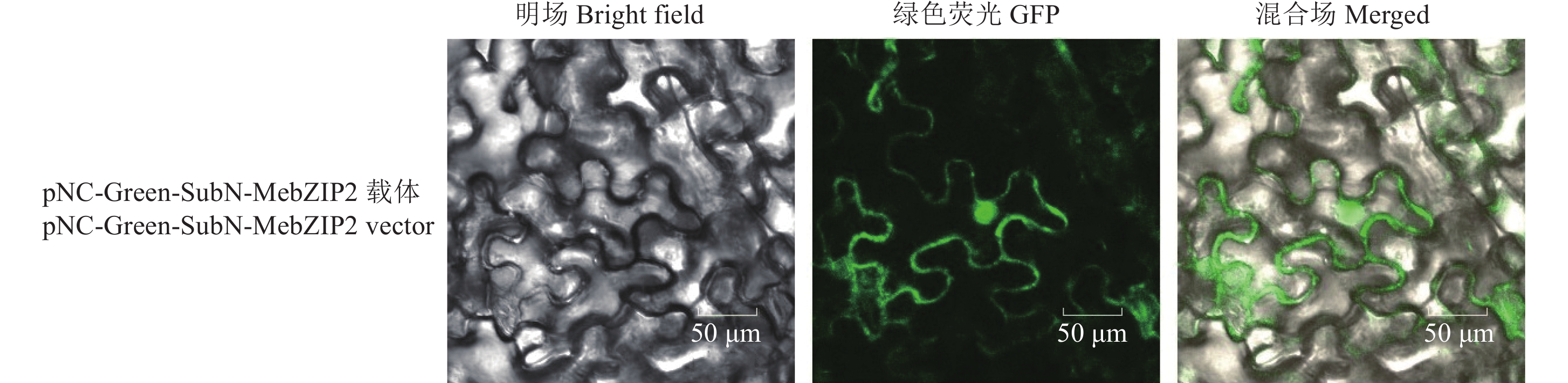
目的 bZIP转录因子在植物生长发育、淀粉合成和非生物胁迫等方面发挥重要作用。克隆木薯MebZIP2基因并进行亚细胞定位,基因表达分析和酵母单杂交分析,为进一步研究MebZIP2基因的功能提供参考。 方法 从木薯SC205中扩增MebZIP2基因编码区(CDS)序列,对其进行生物信息学分析,并构建pNC-Green-SubN融合表达载体,通过农杆菌介导转染烟草表皮细胞,观察荧光信号以确定MebZIP2蛋白的亚细胞定位情况。在木薯不同组织和块根不同发育阶段分析MebZIP2基因表达特征,利用酵母单杂交研究其与淀粉合成基因启动子的互作情况。 结果 MebZIP2基因CDS序列长度为465 bp,编码154个氨基酸,蛋白分子量为17 891.35 Da,理论等电点为5.23,属于不稳定亲水性蛋白。MebZIP2含有bZIP保守结构域,其氨基酸序列与橡胶树bZIP蛋白的相似性最高,为74.22%。MebZIP2基因启动子区域含有光响应元件,赤霉素、水杨酸和茉莉酸等激素响应元件,以及胚乳表达和胁迫响应元件。MebZIP2蛋白亚细胞定位于细胞核和细胞膜中。MebZIP2基因在根尖分生组织、须根、茎和块根发育前期的表达量较高。MebZIP2与淀粉合成基因MeAPL5a、MeGBSS1和MeISA1共表达,而且MebZIP2蛋白可以与它们的启动子互作。 结论 MebZIP2基因属于bZIP基因家族成员,其表达具有组织特异性和块根发育阶段性,MebZIP2蛋白可以与淀粉合成基因MeAPL5a、MeGBSS1和MeISA1的启动子互作,可能参与了木薯块根发育和淀粉合成。 Abstract: :Objective bZIP transcription factors play an important role in plant growth and development, starch synthesis, and abiotic stress. Cloning of cassava MebZIP2 and analysis of its subcellular localization, gene expression, and yeast one-hybrid can provide useful references to further study the function of MebZIP2. Method The coding sequence (CDS) of MebZIP2 was amplified from cassava SC205 and then used to perform bioinformatics analysis. A pNC-Green-SubN fusion expression vector was constructed and transferred into tobacco epidermal cells by Agrobacterium-mediated method, and fluorescence signals were observed to determine subcellular localization of the MebZIP2 protein. The expression profiles of MebZIP2 were analyzed in different tissues and at different development stages of cassava storage root. The interactions between MebZIP2 and the promoters of starch synthesis genes were studied by yeast one-hybrid. Result The CDS sequence of MebZIP2 was 465 bp in length and encoded 154 amino acids. The molecular weight was 17891.35 Da, the theoretical isoelectric point was 5.23, and MebZIP2 was an unstable hydrophilic protein. MebZIP2 contained a bZIP-conserved domain and showed the highest sequence similarity (74.22%) to a bZIP protein from the Hevea brasiliensis. The promoter region of MebZIP2 contained light-responsive elements, hormone (such as gibberellin, salicylic acid, and jasmonic acid) responsive elements, as well as endosperm expression and stress-responsive elements. MebZIP2 protein was localized both in the cell membrane and nucleus. MebZIP2 showed high expression levels in root apical meristems, fibrous roots, stems, and at early developmental stages of storage roots. MebZIP2 was co-expressed with several starch synthesis genes such as MeAPL5a, MeGBSS1, and MeISA1, and MebZIP2 could interact with their promoters. Conclusion MebZIP2 belonged to the bZIP gene family, and its expression showed tissue-specificity and root developmental stage-specificity. MebZIP2 protein could interact with the promoters of starch synthesis genes (MeAPL5a, MeGBSS1 and MeISA1), and it might be involved in storage root development and starch synthesis in cassava. -
Key words:
- Cassava /
- bZIP transcription factor /
- MebZIP2 /
- gene cloning /
- functional analysis
-
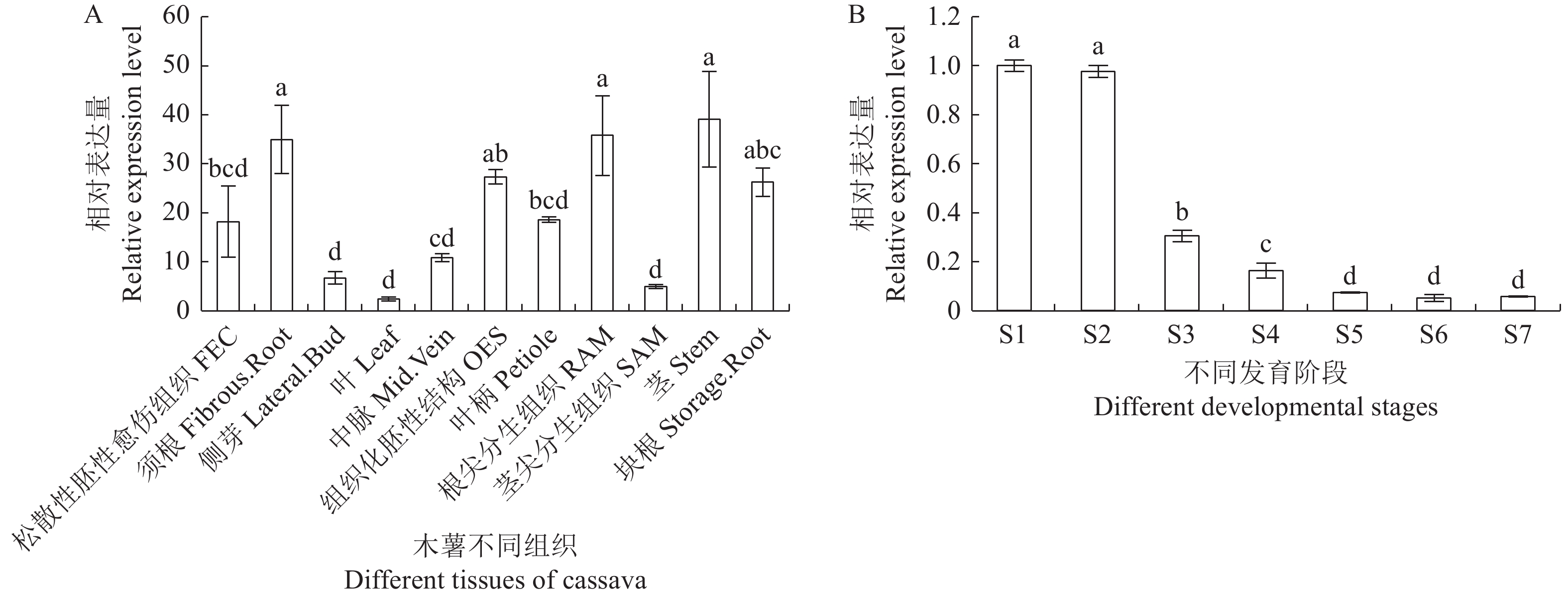
图 6 MebZIP2基因表达分析
(A)MebZIP2在木薯不同组织中的表达分析。(B)MebZIP2在木薯块根不同发育阶段的表达分析。S1~S7表示木薯种植后第100、140、180、220、260、300、340天。不同字母表示均值之间显著差异(邓肯多重检验,P < 0.05)。
Figure 6. MebZIP2 gene expression analysis
(A) Expression analysis of MebZIP2 in different tissues of cassava. (B) Expression analysis of MebZIP2 at different developmental stages of cassava storage root. S1~S7 indicate the 100th, 140th, 180th, 220th, 260th, 300th, 340th day after cassava planting. Different letters indicate significant differences between means (Duncan multiple test, P < 0.05).
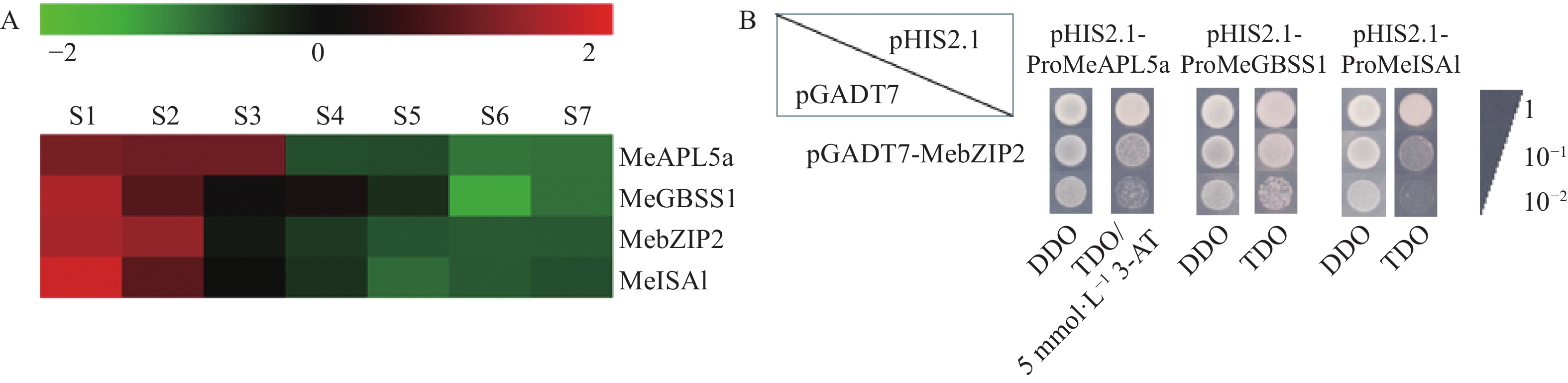
图 7 MebZIP2与淀粉合成基因启动子的互作
A. MebZIP2与淀粉合成基因MeAPL5a、MeGBSS1、MeISA1在木薯块根不同发育阶段的表达趋势。S1~S7表示木薯种植后第100、140、180、220、260、300、340天。B. MebZIP2与淀粉合成基因MeAPL5a、MeGBSS1、MeISA1启动子的酵母单杂交结果。DDO:SD/-Trp-Leu培养基;TDO:不含3-AT的SD/-Trp/-Leu/-His培养基;TDO/5 mmol·L−1 3-AT:含5 mmol·L−1 3-AT的SD/-Trp/-Leu/-His培养基。
Figure 7. Interaction of MebZIP2 with promoters of starch biosynthesis genes
A. Expression analysis of MebZIP2 and starch biosynthesis genes (MeAPL5a, MeGBSS1 and MeISA1) at different developmental stages of cassava storage roots. S1~S7 indicate the 100th, 140th, 180th, 220th, 260th, 300th, 340th day after cassava planting. B. Yeast one-hybrid results between MebZIP2 and the promoters of MeAPL5a, MeGBSS1, and MeISA1, respectively. DDO: SD/-Trp-Leo medium; TDO: SD/-Trp/-Leu/-His medium without 3-AT; TDO/5 mmol·L−1 3-AT: SD/-Trp/-Leu/-His medium with 5 mmol·L−1 3-AT.
表 1 供试引物序列
Table 1. The primers used in this work
引物名称
Primer name引物序列
Primer sequence引物用途
Primer usageMebZIP2-F 5'-GCCATGGAGGCCAGTGAATTCATGGTCTCTTCTAGTGGGGCAT-3′ 基因克隆 MebZIP2-R 5′-CAGCTCGAGCTCGATGGATCCGAACAGGGCCATGTCCATG-3′ pNC-MebZIP2-F 5′-AGTGGTCTCTGTCCAGTCCTATGGTCTCTTCTAGTGGGGCAT-3′ 亚细胞定位 pNC-MebZIP2-R 5′-GGTCTCAGCAGACCACAAGTGAACAGGGCCATGTCCATG-3′ qRTMebZIP2-F 5′-GGATCAGCTCGAGAAAGAAAAC-3′ 荧光定量PCR qRTMebZIP2-R 5′-TTAGGCCATTGCTGAAATTCAC-3′ Actin-F 5′-TGATGAGTCTGGTCCATCCA-3′ 内参基因 Actin-R 5′-CCTCCTACGACCCAATCTCA-3′ MeAPL5a-F 5′-GACTCACTATAGGGCGAATTCGGAAGTGAGCAATAAAAGTAGAAATG-3′ 启动子克隆 MeAPL5a-R 5′-ATAATGCCAGGAATTACTAGTAGCAGCTGAATGTGGAGTGTTT-3′ MeGBSSI-F 5′-GACTCACTATAGGGCGAATTCGAAATGTAGGACCCTGGCGA-3′ 启动子克隆 MeGBSSI-R 5′-ATAATGCCAGGAATTACTAGTCTCTATGCGGAAACAATTAAATCTG-3′ MeISA1-F 5′-GACTCACTATAGGGCGAATTCCTTGAAAATGAAAAAGATTAAAGAGAAA-3′ 启动子克隆 MeISA1-R 5′-ATAATGCCAGGAATTACTAGTCGTTGGTATTACTTGGTGATAATCAC-3′ -
[1] 唐杰, 李明娟, 张雅媛, 等. 食用木薯的加工现状及发展前景 [J]. 食品工业科技, 2023, 44(2):469−476.TANG J, LI M J, ZHANG Y Y, et al. Processing utilization and development prospect of edible cassava [J]. Science and Technology of Food Industry, 2023, 44(2): 469−476. (in Chinese) [2] 付海天, 郑华, 文峰, 等. 中国木薯研究及产业发展趋势 [J]. 农业研究与应用, 2022, 35(4):9−22. doi: 10.3969/j.issn.2095-0764.2022.04.003FU H T, ZHENG H, WEN F, et al. Research and industrial development of cassava in China [J]. Agricultural Research and Application, 2022, 35(4): 9−22. (in Chinese) doi: 10.3969/j.issn.2095-0764.2022.04.003 [3] 曹升, 尚小红, 陈会鲜, 等. 广西地方面包木薯种质资源调查及表型性状分析和品质评价 [J]. 西南农业学报, 2021, 34(11):2318−2325.CAO S, SHANG X H, CHEN H X, et al. Investigation and collection of local bread-cassava germplasm resources in Guangxi and their phenotypic trait analysis and quality evaluation [J]. Southwest China Journal of Agricultural Sciences, 2021, 34(11): 2318−2325. (in Chinese) [4] 蒋瑶, 胡尚连, 孙霞, 等. 拟南芥bZIP基因家族系统发育树比较分析 [J]. 华北农学报, 2008, 23(1):22−27. doi: 10.7668/hbnxb.2008.01.005JIANG Y, HU S L, SUN X, et al. Comparative phylogenetic analysis of the bZIP gene family in Arabidopsis thaliana [J]. Acta Agriculturae Boreali-Sinica, 2008, 23(1): 22−27. (in Chinese) doi: 10.7668/hbnxb.2008.01.005 [5] 韩聪, 何禹畅, 吴丽娟, 等. 水稻碱性亮氨酸拉链(bZIP)蛋白家族功能研究进展 [J]. 中国水稻科学, 2023, 37(4):436−448.HAN C, HE Y C, WU L J, et al. Research progress in the function of basic leucine zipper(bZIP)protein family in rice [J]. Chinese Journal of Rice Science, 2023, 37(4): 436−448. (in Chinese) [6] 董庆. 玉米淀粉合成调控基因的挖掘及功能分析[D]. 合肥: 安徽农业大学, 2015.DONG Q. Identification and characterization of regulatory genes related to starch synthesis in maize(Zea may L. )[D]. Hefei: Anhui Agricultural University, 2015. (in Chinese) [7] E Z G, ZHANG Y P, ZHOU J H, et al. Mini review roles of the bZIP gene family in rice [J]. Genetics and Molecular Research, 2014, 13(2): 3025−3036. doi: 10.4238/2014.April.16.11 [8] DRÖGE-LASER W, SNOEK B L, SNEL B, et al. The Arabidopsis bZIP transcription factor family—An update [J]. Current Opinion in Plant Biology, 2018, 45: 36−49. doi: 10.1016/j.pbi.2018.05.001 [9] ZHANG B Y, FENG C, CHEN L, et al. Identification and functional analysis of bZIP genes in cotton response to drought stress [J]. International Journal of Molecular Sciences, 2022, 23(23): 14894. doi: 10.3390/ijms232314894 [10] WEI K F, CHEN J, WANG Y M, et al. Genome-wide analysis of bZIP-encoding genes in maize [J]. DNA Research, 2012, 19(6): 463−476. doi: 10.1093/dnares/dss026 [11] BI C X, YU Y H, DONG C H, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat [J]. Plant Biotechnology Journal, 2021, 19(2): 209−211. doi: 10.1111/pbi.13453 [12] YUE L, PEI X X, KONG F J, et al. Divergence of functions and expression patterns of soybean bZIP transcription factors [J]. Frontiers in Plant Science, 2023, 14: 1150363. doi: 10.3389/fpls.2023.1150363 [13] WANG Q, GUO C, LI Z Y, et al. Identification and analysis of bZIP family genes in potato and their potential roles in stress responses [J]. Frontiers in Plant Science, 2021, 12: 637343. doi: 10.3389/fpls.2021.637343 [14] 刘西西. SAPK蛋白与bZIP蛋白互作组分析及SAPK10基因调控水稻抽穗期的功能研究[D]. 北京: 中国农业科学院, 2019LIU X X. Analysis of interactions between SAPKs and bZIPs and functional characterization of SAPK10 gene in rice flowering[D]. Beijing: Chinese Academy of Agricultural Sciences, 2019. (in Chinese) [15] NORÉN LINDBÄCK L, JI Y, CERVELA-CARDONA L, et al. An interplay between bZIP16, bZIP68, and GBF1 regulates nuclear photosynthetic genes during photomorphogenesis in Arabidopsis [J]. The New Phytologist, 2023, 240(3): 1082−1096. doi: 10.1111/nph.19219 [16] KUMAR P, KUMAR P, SHARMA D, et al. Genome-wide identification and expression profiling of basic leucine zipper transcription factors following abiotic stresses in potato ( Solanum tuberosum L. ) [J]. PLoS One, 2021, 16(3): e0247864. doi: 10.1371/journal.pone.0247864 [17] 黄星群. 油菜种子贮藏物质合成及外源激素调控的生化分析[D]. 长沙: 湖南大学, 2015.HUANG X Q. Biochemical analysis of storage substances biosynthesis and the regulation of exogenous hormones in Brassica napus L[D]. Changsha: Hunan University, 2015. (in Chinese) [18] DASH M, YORDANOV Y S, GEORGIEVA T, et al. Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress [J]. The Plant Journal, 2017, 89(4): 692−705. doi: 10.1111/tpj.13413 [19] MA H Z, LIU C, LI Z X, et al. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development [J]. Plant Physiology, 2018, 178(2): 753−770. doi: 10.1104/pp.18.00436 [20] DONG Q, XU Q Q, KONG J J, et al. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice [J]. Plant Science, 2019, 283: 407−415. doi: 10.1016/j.plantsci.2019.03.001 [21] KUMAR P, PARVEEN A, SHARMA H, et al. Understanding the regulatory relationship of abscisic acid and bZIP transcription factors towards amylose biosynthesis in wheat [J]. Molecular Biology Reports, 2021, 48(3): 2473−2483. doi: 10.1007/s11033-021-06282-4 [22] WANG L, CAO H L, QIAN W J, et al. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic Arabidopsis [J]. Annals of Botany, 2017, 119(7): 1195−1209. doi: 10.1093/aob/mcx011 [23] WILSON M C, MUTKA A M, HUMMEL A W, et al. Gene expression atlas for the food security crop cassava [J]. The New Phytologist, 2017, 213(4): 1632−1641. doi: 10.1111/nph.14443 [24] FU L L, DING Z H, TIE W W, et al. Integrated metabolomic and transcriptomic analyses reveal novel insights of anthocyanin biosynthesis on color formation in cassava tuberous roots [J]. Frontiers in Nutrition, 2022, 9: 842693. doi: 10.3389/fnut.2022.842693 [25] 颜彦, 铁韦韦, 丁泽红, 等. 木薯MePYL8基因克隆及表达分析 [J]. 分子植物育种, 2018, 16(14):4498−4504.YAN Y, TIE W W, DING Z H, et al. Cloning and expression analysis of MePYL8 gene in cassava [J]. Molecular Plant Breeding, 2018, 16(14): 4498−4504. (in Chinese) [26] DING Z H, FU L L, TIE W W, et al. Highly dynamic, coordinated, and stage-specific profiles are revealed by a multi-omics integrative analysis during tuberous root development in cassava [J]. Journal of Experimental Botany, 2020, 71(22): 7003−7017. doi: 10.1093/jxb/eraa369 [27] 张开慧. 酵母单杂交技术的原理及应用 [J]. 宁夏农林科技, 2012, 53(2):31−32. doi: 10.3969/j.issn.1002-204X.2012.02.016ZHANG K H. Principle and application of yeast one-hybrid method [J]. Ningxia Journal of Agriculture and Forestry Science and Technology, 2012, 53(2): 31−32. (in Chinese) doi: 10.3969/j.issn.1002-204X.2012.02.016 [28] EDRISI MARYAN K, FARROKHI N, SAMIZADEH LAHIJI H. Cold-responsive transcription factors in Arabidopsis and rice: A regulatory network analysis using array data and gene co-expression network [J]. PLoS One, 2023, 18(6): e0286324. doi: 10.1371/journal.pone.0286324 [29] WANG J C, XU H, ZHU Y, et al. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm [J]. Journal of Experimental Botany, 2013, 64(11): 3453−3466. doi: 10.1093/jxb/ert187 [30] SONG Y H, LUO G B, SHEN L S, et al. TubZIP28, a novel bZIP family transcription factor from Triticum urartu, and TabZIP28, its homologue from Triticum aestivum, enhance starch synthesis in wheat [J]. The New Phytologist, 2020, 226(5): 1384−1398. doi: 10.1111/nph.16435 [31] PONTES L C G, CARDOSO C M Y, CALLEGARI D M, et al. A cassava CPRF-2-like bZIP transcription factor showed increased transcript levels during light treatment [J]. Protein and Peptide Letters, 2020, 27(9): 904−914. doi: 10.2174/0929866527666200420110338 [32] ZHANG Z R, QUAN S W, NIU J X, et al. Genome-wide identification, classification, expression and duplication analysis of bZIP family genes in Juglans regia L [J]. International Journal of Molecular Sciences, 2022, 23(11): 5961. doi: 10.3390/ijms23115961 [33] LIU H T, TANG X, ZHANG N, et al. Role of bZIP transcription factors in plant salt stress [J]. International Journal of Molecular Sciences, 2023, 24(9): 7893. doi: 10.3390/ijms24097893 [34] ZHU Q Y, LV J H, WU Y, et al. MdbZIP74 negatively regulates osmotic tolerance and adaptability to moderate drought conditions of apple plants [J]. Journal of Plant Physiology, 2023, 283: 153965. doi: 10.1016/j.jplph.2023.153965 [35] CAI J, XUE J J, ZHU W L, et al. Integrated metabolomic and transcriptomic analyses reveals sugar transport and starch accumulation in two specific germplasms of Manihot esculenta crantz [J]. International Journal of Molecular Sciences, 2023, 24(8): 7236. doi: 10.3390/ijms24087236 [36] 未丽, 刘建利. 植物蛋白质亚细胞定位相关研究概述 [J]. 植物科学学报, 2021, 39(1):93−101. doi: 10.11913/PSJ.2095-0837.2021.10093WEI L, LIU J L. Overview of research on protein subcellular localization in plants [J]. Plant Science Journal, 2021, 39(1): 93−101. (in Chinese) doi: 10.11913/PSJ.2095-0837.2021.10093 [37] PAN X J, WANG C L, LIU Z S, et al. Identification of ABF/ AREB gene family in tomato ( Solanum lycopersicum L. ) and functional analysis of ABF/ AREB in response to ABA and abiotic stresses [J]. PeerJ, 2023, 11: e15310. doi: 10.7717/peerj.15310 [38] SONG M, FANG S Q, LI Z G, et al. CsAtf1, a bZIP transcription factor, is involved in fludioxonil sensitivity and virulence in the rubber tree anthracnose fungus Colletotrichum siamense [J]. Fungal Genetics and Biology, 2022, 158: 103649. doi: 10.1016/j.fgb.2021.103649 [39] WANG B X, XU B, LIU Y, et al. A Novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice ( Oryza sativa L. ) [J]. Plant Science, 2022, 323: 111393. doi: 10.1016/j.plantsci.2022.111393 [40] FU L L, DING Z H, TIE W W, et al. Large-scale RNAseq analysis reveals new insights into the key genes and regulatory networks of anthocyanin biosynthesis during development and stress in cassava [J]. Industrial Crops and Products, 2021, 169: 113627. doi: 10.1016/j.indcrop.2021.113627 -








 下载:
下载: