Cloning and expression analysis of RhMAX2A gene in Rosa hybrid
-
摘要:
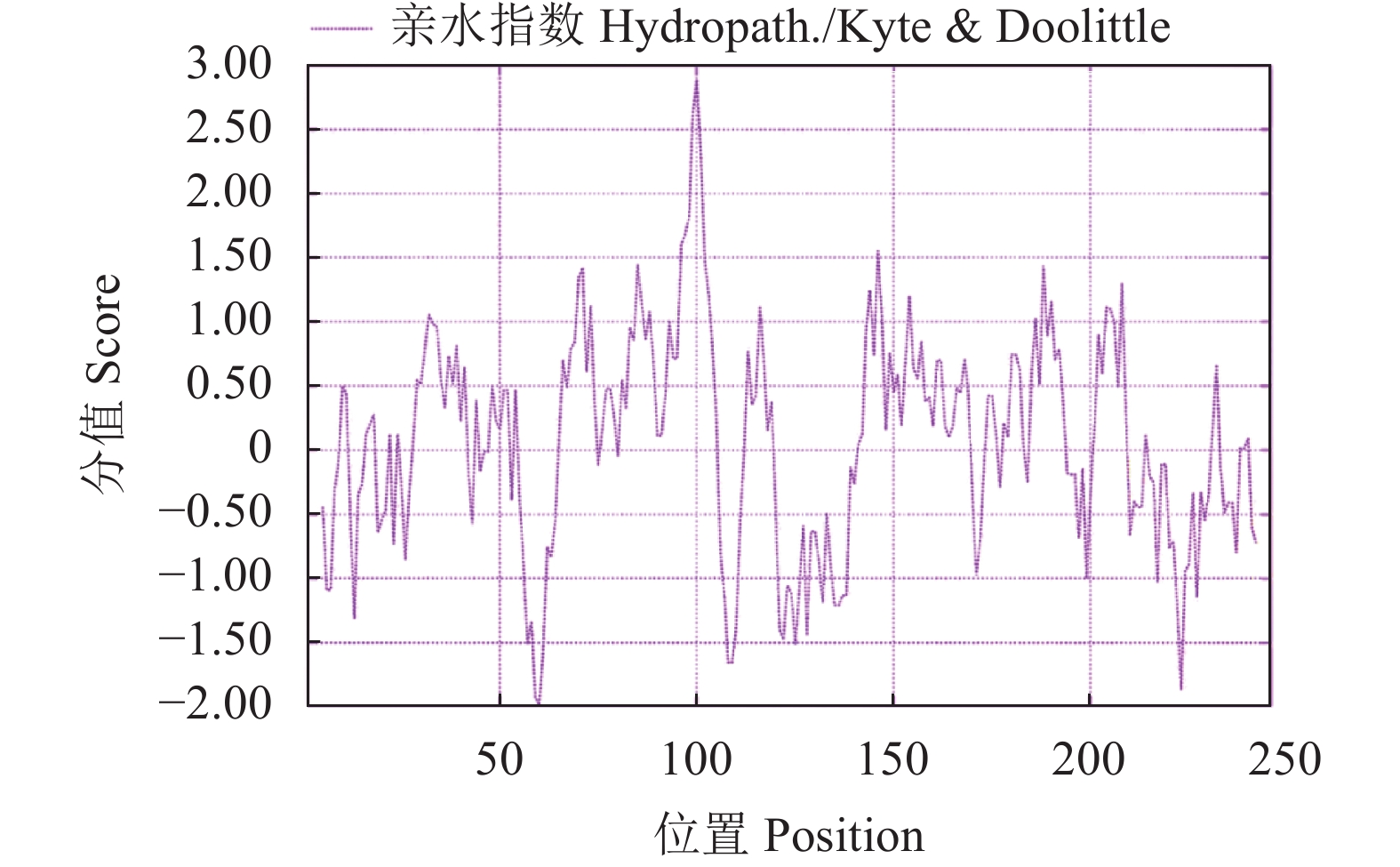
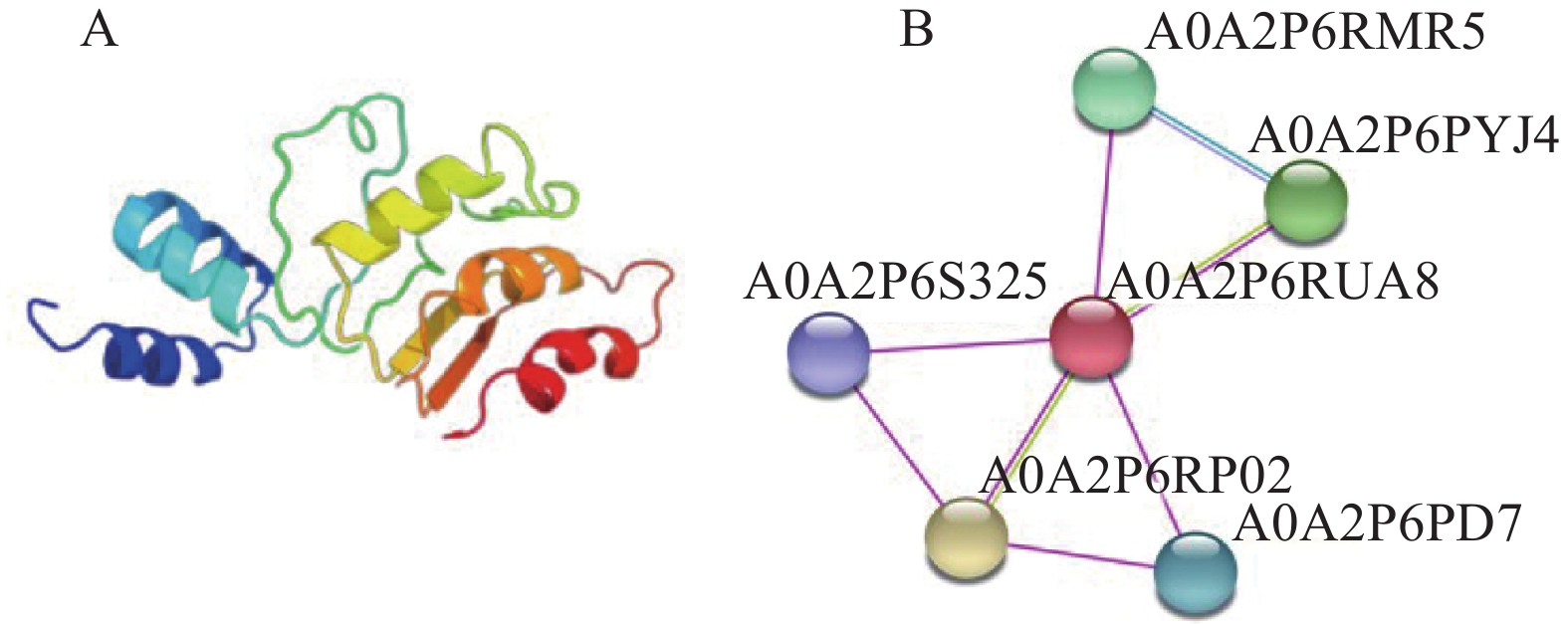
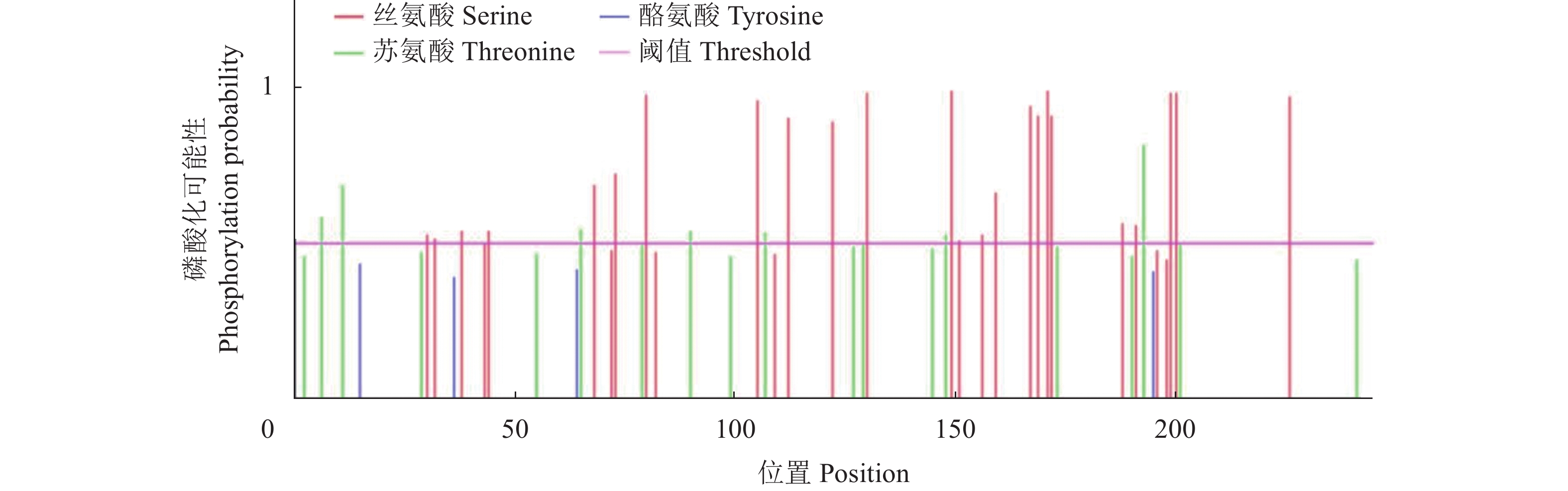
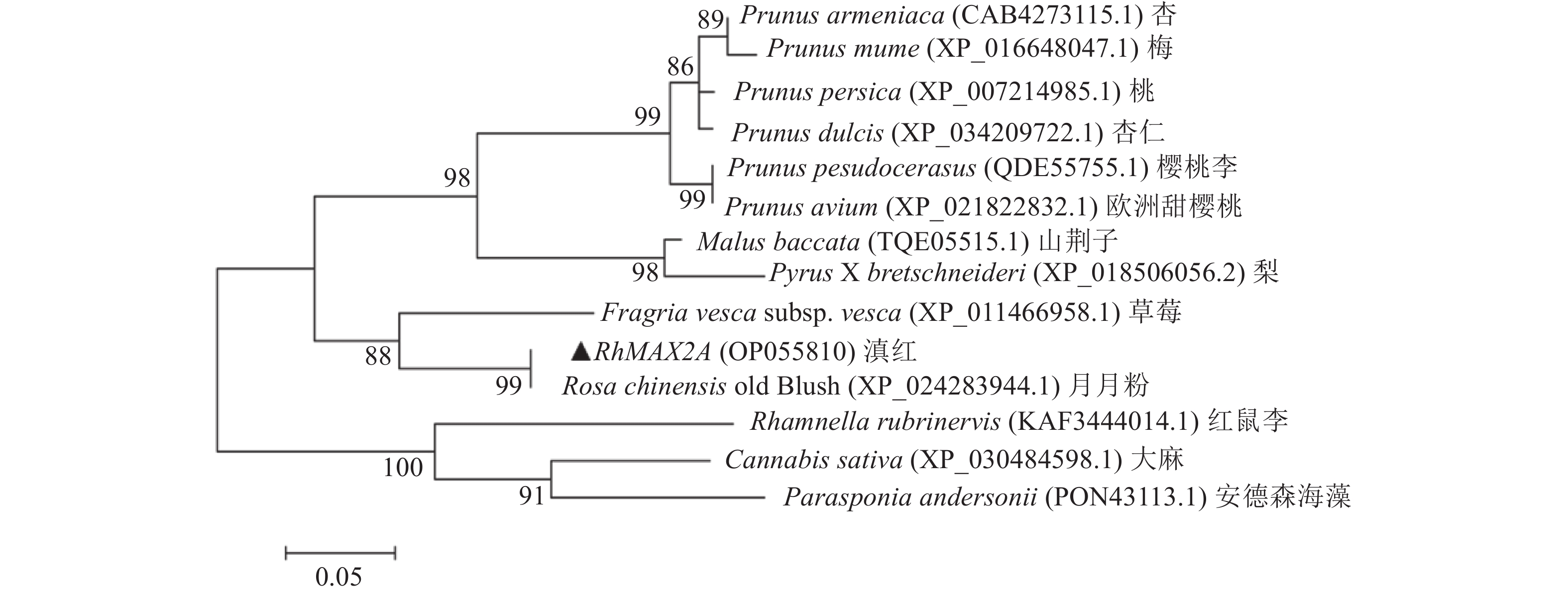
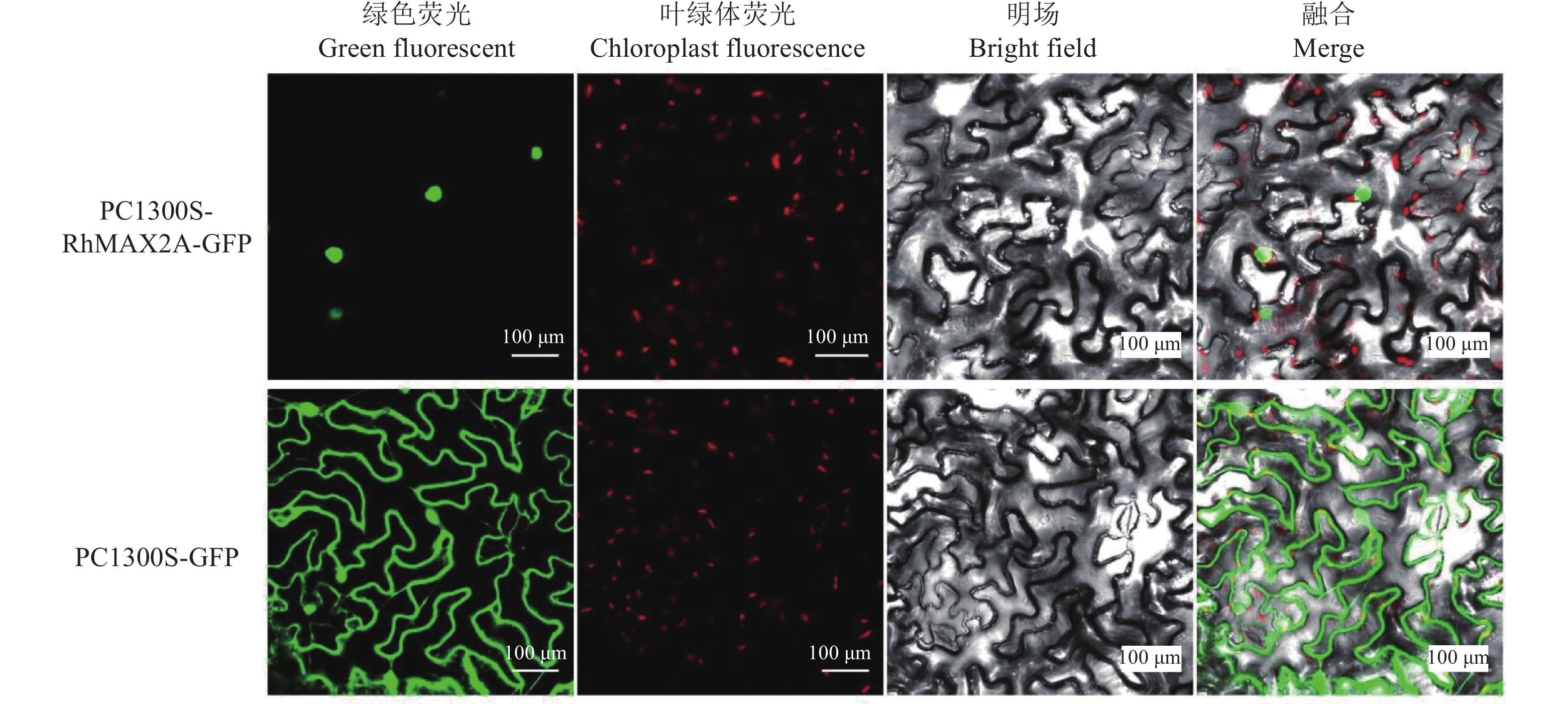
目的 克隆月季RhMAX2A基因cDNA序列,并分析其序列特征及其在不同组织中和去顶后的表达情况,为探究该基因在月季中的生物学功能及调控侧枝发生的转导机制提供理论支持。 方法 以月季品种滇红为材料,通过 RT-PCR 技术克隆RhMAX2基因的cDNA序列,利用生物信息学方法对其序列和所编码的蛋白质进行分析,利用烟草瞬时转化技术分析蛋白的亚细胞定位,同时采用实时荧光定量PCR(qRT-PCR)检测其在不同组织中及去顶后的表达情况。 结果 RhMAX2A基因(GeneBank登录号为OP055810)cDNA序列长1030 bp,编码246 个氨基酸,该蛋白分子式为C2910H4793N1029O1244S210,相对分子质量为27.35 kD,总原子量为3909;该蛋白不稳定系数为53.07,脂肪系数为106.30,GRAVY值为0.049,是一类不稳定亲水性蛋白;RhMAX2A蛋白的二级结构主要由α-螺旋和无规则卷曲构成,且RhMAX2A为推定的F-box结构域,属于α/β水解酶家族;同源序列比对和系统进化树关系分析结果表明,RhMAX2A 氨基酸序列(OP055810)与同属的古老月季品种月月粉氨基酸序列(XP_024283944.1)相似性最高,其次是同亚科的草莓(XP_004287076.1),三者亲缘关系较近。亚细胞定位结果显示,RhMAX2A编码蛋白位于细胞核。qRT-PCR检测结果显示,RhMAX2A基因在根、腋芽和节中表达,根中表达量最高,去顶处理显著上调RhMAX2A基因在根和腋芽中的表达。 结论 成功克隆了滇红RhMAX2A基因,其编码蛋白在细胞核上发挥作用,主要在根和腋芽中表达,且受去顶诱导上调表达。 Abstract: :Objective To study the biological function and the transduction mechanism that regulates lateral branching of the RhMAX2A gene in Rosa, the cDNA sequence of RhMAX2A was cloned and was analysed for sequence characteristics and its expression in different tissues and after decapitation. Methods The cDNA sequence of RhMAX2A gene was cloned from Rosa hybrida ‘Dianhong’ by RT-PCR and the bioinformatics analysis was conducted. The subcellular location of RhMAX2A was analyzed by transient transformation of PC1300s-RhMAX2A-GFP in tobacco leaves. The expression levels of RhMAX2A in different tissues and the treatment of decapitation was detected by real-time quantitative PCR (qRT-PCR). Results The cDNA sequence of RhMAX2A gene (GenBank accession number: OP055810 ) was 1030 bp in length, encoding 246 amino acids. The calculated chemical molecular formula of RhMAX2A protein is C2910H4793N1029O1244S210, with 27.35 kD in molecular mass and the total atomic weight is 3909. The instability coefficient of this protein is 53.07, with fat coefficient-106.30 and GRAVY value 0.049, indicating that it is an unstable hydrophilic protein. The secondary structure of RhMAX2A protein is mainly composed of α-helix and random coil, and RhMAX2A is a presumed F-box domain, belonging to the α/β hydrolase family. Homologous sequence alignment and phylogenetic tree analysis showed that the amino acid sequence of RhMAX2A (OP055810) had the highest similarity with that of Rosa chinensis Old Blush (XP_024283944.1), followed by Fragaria vesca subsp. vesca (XP_004287076.1) of the same subfamily, which were closely related. The subcellular localization results showed that the encoded protein of RhMAX2A was located in the nucleus. The results of qRT-PCR indicated that RhMAX2A gene was expressed in roots, axillary buds and nodes, with the highest expression level in roots. The decapitation treatment significantly upregulated the expression of RhMAX2A in roots and axillary buds. Conclusion The RhMAX2A gene was successfully cloned from Rosa hybrida ‘Dianhong’, which encodes a protein that plays a role in the nucleus, mainly expressed in roots and axillary buds, and could be upregulated by decapitation. -
Key words:
- Rosa hybrida /
- MAX2A gene /
- gene cloning /
- subcellular localization /
- expression analysis
-
图 13 RhMAX2A基因在不同组织及去顶处理下的表达模式
A. 不同组织,B. 根,C. 腋芽; ns表示无显著差异(P > 0.05),*表示差异显著(P < 0.05),**表示差异极显著(P < 0.01),***表示差异极显著(P < 0.001)。
Figure 13. Expression patterns of RhMAX2A in different tissues and under decapitation
A. Different tissues, B. Roots, C. Axillary buds; ns indicates no significant (P > 0.05), * indicates significant difference (P < 0.05), ** indicates extremely significant difference (P < 0.01), *** indicates extremely significant difference (P < 0.001).
表 1 引物序列
Table 1. Primer sequences
引物名称 Primer 引物序列 Primer sequence RhMAX2A-F 5′-TCTCTCTCGAGCTTTCGCGAGCTCATGCAATCGCGTTCACGTG-3′ RhMAX2A-R 5′-TCGCCCTTGCTCACCATGGATCCATCAAGGATTGTGCGCCTGT-3′ 18S rRNA-F 5′-CCTGAGAAACGGCTACCACAT-3′ 18S rRNA-R 5′-CACCAGACTTGCCCTCCA-3′ UBC-F 5′-GCCAGAGATTGCCCATATGTGTA-3′ UBC-R 5′-TCACAGAGTCCTAGCAGCACA-3′ RhMAX2A-qRT-F 5′-GATCGACTTCTTCTCCGGGCTT-3′ RhMAX2A-qRT-R 5′-CCAATCCACTGTCCCTCACGTT-3′ RhMAX2A-GFP-F 5′-TCTCTCTCGAGCTTTCGCGAGCTCATGCAATCGCGTTCACGTG-3′ RhMAX2A-GFP-R 5′-TCGCCCTTGCTCACCATGGATCCATCAAGGATTGTGCGCCTGT-3′ -
[1] 王榕, 牛晓茹, 陈己任. 月季 RcTCP20基因的克隆及在拟南芥中的功能分析 [J]. 植物生理学报, 2022, 58(10):1907−1918.WANG R, NIU X R, CHEN J R. Cloning of RcTCP20 of rose and function analysis in Arabidopsis thaliana [J]. Plant Physiology Journal, 2022, 58(10): 1907−1918. [2] YOUNG E L, WU X, LIANG S, et al. Heritability of plant architecture in diploid roses [J]. Acta Horticulturae, 2019(1232): 15−18. [3] KAWAMURA K, HIBRAND-SAINT OYANT L, CRESPEL L, et al. Quantitative trait loci for flowering time and inflorescence architecture in rose [J]. Theoretical and Applied Genetics, 2011, 122(4): 661−675. doi: 10.1007/s00122-010-1476-5 [4] AL-BABILI S, BOUWMEESTER H J. Strigolactones, a novel carotenoid-derived plant hormone [J]. Annual Review of Plant Biology, 2015, 66: 161−186. doi: 10.1146/annurev-arplant-043014-114759 [5] 杜高齐, 李雪娇, 许彬, 等. 月季 RhD14基因克隆、亚细胞定位及表达分析 [J]. 南方农业学报, 2023, 54(1):34−45.DU G Q, LI X J, XU B, et al. Cloning, subcellular localization and expression analysis of RhD14 gene in rose [J]. Journal of Southern Agriculture, 2023, 54(1): 34−45. [6] KONG X P, ZHANG M L, DING Z J. D53: The missing link in strigolactone signaling [J]. Molecular Plant, 2014, 7(5): 761−763. doi: 10.1093/mp/ssu016 [7] STRUK S, DE CUYPER C, JACOBS A, et al. Unraveling the MAX2 protein network in Arabidopsis thaliana: Identification of the protein phosphatase PAPP5 as a novel MAX2 interactor [J]. Molecular & Cellular Proteomics:MCP, 2021, 20: 100040. [8] WATERS M T, GUTJAHR C, BENNETT T, et al. Strigolactone signaling and evolution [J]. Annual Review of Plant Biology, 2017, 68: 291−322. doi: 10.1146/annurev-arplant-042916-040925 [9] SHEN H, LUONG P, HUQ E. The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis [J]. Plant Physiology, 2007, 145(4): 1471−1483. doi: 10.1104/pp.107.107227 [10] YAO R F, MING Z H, YAN L M, et al. DWARF14 is a non-canonical hormone receptor for strigolactone [J]. Nature, 2016, 536: 469−473. doi: 10.1038/nature19073 [11] WANG Y, SUN S Y, ZHU W J, et al. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching [J]. Developmental Cell, 2013, 27(6): 681−688. doi: 10.1016/j.devcel.2013.11.010 [12] WANG L, WANG B, JIANG L, et al. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation [J]. The Plant Cell, 2015, 27(11): 3128−3142. doi: 10.1105/tpc.15.00605 [13] ZHOU F, LIN Q B, ZHU L H, et al. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling [J]. Nature, 2013, 504: 406−410. doi: 10.1038/nature12878 [14] DONG L L, ISHAK A, YU J, et al. Identification and functional analysis of three MAX2 orthologs in chrysanthemum [J]. Journal of Integrative Plant Biology, 2013, 55(5): 434−442. doi: 10.1111/jipb.12028 [15] ZHAO L L, FANG J J, XING J, et al. Identification and functional analysis of two cotton orthologs of MAX2 which control shoot lateral branching [J]. Plant Molecular Biology Reporter, 2017, 35(5): 480−490. doi: 10.1007/s11105-017-1040-4 [16] 苏倩, 杜文宣, 马琳, 等. 紫花苜蓿MsCIPK2的克隆及功能分析 [J]. 中国农业科学, 2022, 55(19):3697−3709. doi: 10.3864/j.issn.0578-1752.2022.19.002SU Q, DU W X, MA L, et al. Cloning and functional analyses of MsCIPK2 in Medicago sativa [J]. Scientia Agricultura Sinica, 2022, 55(19): 3697−3709. (in Chinese) doi: 10.3864/j.issn.0578-1752.2022.19.002 [17] 董雪. 瓜列当独脚金内酯感知基因PaMAX2克隆及其功能研究[D]. 石河子: 石河子大学, 2021.DONG X. Cloning and functional study of strigolactone perception gene PaMAX2 in guadalajaran[D]. Shihezi: Shihezi University, 2021. (in Chinese) [18] HE P, ZHANG H Z, ZHANG L, et al. The GhMAX2 gene regulates plant growth and fiber development in cotton [J]. Journal of Integrative Agriculture, 2022, 21(6): 1563−1575. doi: 10.1016/S2095-3119(21)63603-1 [19] 姜思思. 杨树中MAX2同源基因的克隆[D]. 武汉: 华中农业大学, 2016.JIANG S S. Cloning of MAX2 homologous genes in Populus[D]. Wuhan: Huazhong Agricultural University, 2016. (in Chinese) [20] 李国防. 苹果MAX2基因介导独脚金内酯信号调控腋芽萌发的功能研究[D]. 杨凌: 西北农林科技大学, 2018.LI G F. Functional study of MAX2 gene on the regulation of axillary bud outgrowth by mediating strigolactone signaling in Malus[D]. Yangling: Northwest A & F University, 2018. (in Chinese) [21] 朱思雅, 李雪娇, 赵雁, 等. 去顶诱导的食用玫瑰‘滇红’腋芽萌发及内源激素含量变化[J/OL]. 分子植物育种, 1-12[2024-02-29]. http://kns.cnki.net/kcms/detail/46.1068.S.20220804.1416.006.html.ZHU S Y, LI X J, ZHAO Y, et al. Changes of axillary bud outgrowth and endogenous hormone contents of edible rose, R. chinensis Dianhong, induced by decapitation[J]. Molecular Plant Breeding, 1-12[2024-02-29]. http://kns.cnki.net/kcms/detail/46.1068.S.20220804.1416.006.html. [22] 朱思雅. 去顶诱导的月季腋芽形态、内源激素变化及转录组分析[D]. 昆明: 云南农业大学, 2021.ZHU S Y. Axillary bud morphology, endogenous hormone changes and transcriptome analysis of Rose axillary buds induced by topping[D]. Kunming: Yunnan Agricultural University, 2021. (in Chinese) [23] DUN E A, DE SAINT GERMAIN A, RAMEAU C, et al. Dynamics of strigolactone function and shoot branching responses in Pisum sativum [J]. Molecular Plant, 2013, 6(1): 128−140. doi: 10.1093/mp/sss131 [24] TANG Y J, LIESCHE J. The molecular mechanism of shade avoidance in crops–How data from Arabidopsis can help to identify targets for increasing yield and biomass production [J]. Journal of Integrative Agriculture, 2017, 16(6): 1244−1255. doi: 10.1016/S2095-3119(16)61434-X [25] 程亭亭. 柳枝稷分蘖调控基因PvMAX2的功能研究[D]. 杨凌: 西北农林科技大学, 2018.CHENG T T. Functional analysis of tillering-related gene PvMAX2 in switchgrass[D]. Yangling: Northwest A & F University, 2018. (in Chinese) [26] 任广悦, 金晓霞, 陈超, 等. 黄瓜独脚金内酯信号转导基因CsMAX2的克隆与表达分析 [J]. 分子植物育种, 2020, 18(16):5237−5246.REN G Y, JIN X X, CHEN C, et al. Cloning and expression analysis of strigolactone signal transduction gene CsMAX2 in cucumber ( Cucumis sativus) [J]. Molecular Plant Breeding, 2020, 18(16): 5237−5246. (in Chinese) [27] FIORILLI V, FORGIA M, DE SAINT GERMAIN A, et al. A structural homologue of the plant receptor D14 mediates responses to strigolactones in the fungal phytopathogen Cryphonectria parasitica [J]. The New Phytologist, 2022, 234(3): 1003−1017. doi: 10.1111/nph.18013 [28] 袁飞荣, 袁耀明, 李智鑫, 等. 枳橙CPMAX2基因的克隆、载体构建及其表达研究 [J]. 园艺学报, 2015, 42(12):2497−2504.YUAN F R, YUAN Y M, LI Z X, et al. Cloning, constructing the over-expression vector and expression analysis of CPMAX2 in transgenic citrange with rolABC genes [J]. Acta Horticulturae Sinica, 2015, 42(12): 2497−2504. (in Chinese) [29] BASIR UL HAQ. MAX基因参与大豆独角金内酯合成和信号传导的功能研究[D]. 武汉: 华中农业大学, 2018: 35-45.BASIR U. Study on the function of MAX gene involved in the synthesis and signal transduction of soybean monogonolide[D]. Wuhan: Huazhong Agricultural University, 2018: 35-45. (in Chinese) [30] DIERCK R, KEYSER E D, RIEK J D, et al. Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in Chrysanthemum [J]. PLoS One, 2016, 11(8): e0161732. doi: 10.1371/journal.pone.0161732 [31] 程方. 苹果独脚金内酯合成基因MdMAX1调控腋芽萌发的功能研究[D]. 杨凌: 西北农林科技大学, 2020CHENG F. Study on the function of apple strigolactone synthetic gene MdMAX1 in regulating axillary bud germination[D]. Yangling: Northwest A & F University, 2020. (in Chinese) -







 下载:
下载: